bioGUI: a universal GUI for CLI applications
With bioGUI developers can easily build a graphical-user-interface (GUI) for their command-line only application. Such a GUI then enables many more people to use their software. Especially in biology, the domain experts are often computer novices and have difficulties using tools written for them by bioinformaticiens/computer scientists.
You can find the corresponding paper published with PeerJ.
Who is bioGUI for?
With bioGUI domain experts, which have no clue about the command line, should be enabled to use highly sophisticated tools written for them. bioGUI specifically aims at Windows users, as Microsoft just introduced the Windows Subsystem for Linux (WSL) with its Bash on Ubuntu on Windows. This system allows the usage of said sophisticated tools on a regular Windows computer as most people have. With bioGUI one also does not need any knowledge about the command line, because the task of executing a given tools becomes a point & click solution.
Where can I get bioGUI?
bioGUI is available for Windows, Mac OS and Linux as prebuilt-binary distribution or source from github releases. The binary distribution (zip-files) are targeted for end-users: prebuilt binaries.
After downloading the zip-archive, please unzip the archive to a location of your preference. Then simply start the executable (bioGUI.exe on Windows, or bioGUI on linux).
On Windows, please follow the steps on how to setup WSL.
Place bioGUI into a location which does not contain spaces in its name, e.g not C:\Program Files\bioGUI ! C:\bioGUI or D:\bioGUI\ is fine though!
On any aptitude supported platform (Windows with WSL, Ubuntu), please download the "First Time Ubuntu/WSL/apt-get Setup" from the list of available templates and install it via Install Template Module (install program: First Time Ubuntu/WSL/apt-get setup). For Mac OS, please execute download and execute First Time Mac OS Setup.
Please also refer to our User Guide.
Requirements
Linux
Ubuntu 18.04 or similar with aptitude (apt) package management.
Windows
Microsoft Windows 10 (build 17763, Oct. 2018 update) with Windows Subsystem for Linux-Feature enabled and the Ubuntu 18.04-App installed.
This page describes how to setup WSL.
Place bioGUI into a location which does not contain spaces in its name, e.g not C:\Program Files\bioGUI ! C:\bioGUI or D:\bioGUI\ is fine though!
If you want to access external drives (USB stick, network drive), you first need to mount this drive into WSL. The WSL Mount Drive install module will install a script which can do this for you. Make sure to save the template and use the Mount Drive (WSL) template to make the drive available to WSL. You need to enter the drive-letter you want to mount (e.g. F), and your sudo password.
Mac OS
bioGUI requires Mac OS X 10.14 Mojave.
Mac OS may bother you about running an app from a non-verified developer.
In the Preferences->Security menu you can tell Mac OS to still run bioGUI.
Please refer to the User Guide for more instructions.
Setup First Time Use
Install and use an install module
I want to know more about available modules
If you want to know which modules are already available, please visit the supporting information website. There you can see a list of available modules, submit your own or request new template. If you have any problem, or you want to request a new template, you may also use the github issue tracker right in this repository!
Developer's Guides
If you are a developer and want to write a bioGUI (install template), there's good news: it's fairly easy.
Please follow this guide for information on how to write the install module. Within the install module you need to embed/write your actual GUI definition. This is explained in the install template example.
From my experience in writing install modules, it is best to copy an existing install template and modify this, for both the install part, as well as the GUI part.
I want to know more about bioGUI
You can find a detailed documentation of bioGUI at readthedocs.
Instructional Videos
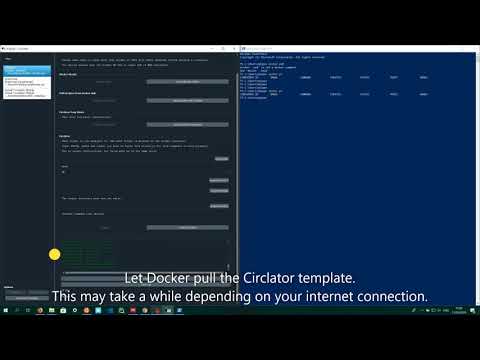
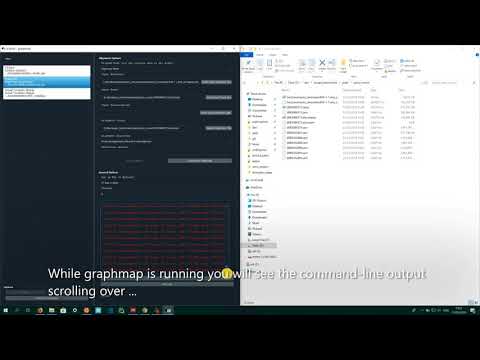
After installing WSL, installing+using graphmap
With a Docker Image
About using/building bioGUI
How to obtain bioGUI?
Currently the only way is to clone this repository and build bioGUI. It is planned to also distribute full installers/archives to download with binaries.
Use releases from GitHub
There are releases built for bioGUI for Linux, Mac OS and Windows.
For Windows (and especially WSL) these are stand-alone.
Under Linux and Mac OS make sure that wgetand nectarare installed.
We recommend to use brew on Mac OS:
brew install wget
brew install netcat
How to build bioGUI?
Before building bioGUI check that the following dependencies are installed:
* cmake
* Qt 5.5+
* libssl-dev
* liblua5.3-dev
then create a new folder build within the git root, cd into it and execute cmake .. && make && make install
How to see which templates exist and how to install new templates?
The Download Templates button in bioGUI lists all available templates and allows also for filtering. You can also explore all templates at our website. This is also where you can submit new/modified templates.
After clicking on Download Templates, you can select one or multiple templates to download. These will then be downloaded and saved for you. Upon exiting the Download Templates dialogue, you will see the templates in bioGUI. Install templates require the Template Installation Module to be downloaded before.
README to be continued
About Templates
Templates are structued XML files.
Each template is defined as a root tag with the following structure:
<template description="a descriptive text" title="a title">
Where the description is shown in the description line in bioGUI and the title is shown in the title line in bioGUI.
If the child element window does not have a title set, the template title is also used as window title.
Within the template tag two child elements are expected:
* `<window width="..." height="..." title="...">`
* `<execution>`
The window may have additional attributes such as width and height defining the minimal width/height of the window. The title is shown in the title of the window.
The window tag (and its children) define the visual appearance of a template while the execution part defines the logical part.
Once starting a program, the execution part can be used to assemble the command line arguments for the application to start.
For this a network-like structure is used.
Each element in the execution part can be seen as a node.
A node can refer its value from another node, but it can also manipulate or extend values (e.g. by combining multiple nodes in an <add>-node).
For highest flexibility, <script> nodes can refer to or contain LUA code. For instance
<const id="node1">some_file.tex</const>
<script argv="${node1},pdf">
<![CDATA[
function evaluate(arg1, arg2)
return(string.sub(arg1, 0, -3) .. arg2)
end
]]>
</script>would first split all supplied arguments from the script argv attribute and resolve those, which refer to another node (indicated by ${nodeid}).
In this case, the node with id node1 is a constant value of some_file.tex. The second argument is also constant text (pdf).
Therefore the inline script would be called as evaluate(some\_file.tex, pdf).
The return value is thus some_file.pdf.
How to create a new template?
In order to create a new template, simply setup a new text file with gui extension (or igui if it's an install template).
Create and set up the visual part of the GUI and write the logic part to assemble the command line arguments.
If you want to share your template, feel free to upload to our website.
Which visual elements exist?
Visual elements including their possible attributes are given below. It is distinguished between Layouts and Widgets. Layouts tell how the widgets are rendered/displayed (e.g. in which order). There currently exist 3 layouts (grid, horizontal and vertical).
| node name | allowed attributes |
|---|---|
| <GRID> | [cols, rows] |
| <HGROUP> | [] |
| <VGROUP> | [] |
| --- | --- |
| <ACTION> | [program] |
| <CHECKBOX> | [selected, selectonwindows, value] |
| <COMBOBOX> | [selected] |
| <COMBOITEM> | [value] |
| <FILEDIALOG> | [filter, folder, location, multiples, multiples_delim, output] |
| <FILELIST> | [height, title, width] |
| <FILESELECTBOX> | [delim, filter, location] |
| <GROUP> | [height, title, width] |
| <GROUPBOX> | [multi] |
| <IMAGE> | [height, src, width] |
| <INPUT> | [multi, type {string, int, float, password}] |
| <LABEL> | [link] |
| <RADIOBUTTON> | [value] |
| <SLIDER> | [max, min, step] |
| <SLIDERITEM> | [display, value] |
| <STREAM> | [height, title, width] |
| <STREAMBOX> | [] |
| <WINDOW> | [height, title, width] |
Which execution nodes exist?
Execution nodes define the logic and how the command line arguments are assembled. Most likely the most important nodes are the add, const, value and if (and else) nodes.
However, it must be stressed, that the script node is of high power as it enables the execution of lua scripts (even inline written in the template). Program arguments are provided by the argv attribute (see the above example).
| node name | allowed attributes |
|---|---|
| <add> | [ID, TYPE, sep] |
| <const> | [ID, TYPE] |
| <else> | [] |
| <env> | [GET, ID, TYPE] |
| <execute> | [EXEC, ID, PROGRAM, TYPE, location, param, program, wsl] |
| <file> | [FROM, ID, SEP, TO, TYPE] |
| <httpexecute> | [CL_TO_POST, DELIM, ID, PORT, PROGRAM, TYPE] |
| <if> | [COMP, ID, SEP, TYPE, VALUE1, VALUE2] |
| <math> | [ID, OP, TYPE] |
| <orderedadd> | [FROM, ID, SELECTED, TYPE] |
| <output> | [COLOR, DEFERRED, FROM, HOST, ID, LOCATION, PORT, TO, TYPE, TYPE] |
| <relocate> | [FROM, ID, PREPEND, TO, TYPE, UNIX, WSL] |
| <replace> | [ID, REPLACE, REPLACE_WITH, TYPE] |
| <script> | [ARGV, ID, SCRIPT, TYPE] |
| <value> | [FOR, FROM, ID, TYPE] |
Licenses
bioGUI
LUA
This project requires a static LUA library to build. For the release of this software, LuaBinaries static liblua53.a was used and requires the following copyright notice to be included:
Copyright © 2005-2016 Tecgraf/PUC-Rio and the Kepler Project.
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
OpenSSL
This project also requires a static OpenSSL library to build. For the release of this software, OpenSSL static libraries were used and require the following copyright notice to be included:
This product includes software developed by the OpenSSL Project for use in the OpenSSL Toolkit (http://www.openssl.org/) This product includes cryptographic software written by Eric Young ([email protected]). This product includes software written by Tim Hudson ([email protected]).
LICENSE ISSUES
==============
The OpenSSL toolkit stays under a dual license, i.e. both the conditions of
the OpenSSL License and the original SSLeay license apply to the toolkit.
See below for the actual license texts.
OpenSSL License
---------------
====================================================================
Copyright (c) 1998-2016 The OpenSSL Project. All rights reserved.
Redistribution and use in source and binary forms, with or without
modification, are permitted provided that the following conditions
are met:
1. Redistributions of source code must retain the above copyright
notice, this list of conditions and the following disclaimer.
2. Redistributions in binary form must reproduce the above copyright
notice, this list of conditions and the following disclaimer in
the documentation and/or other materials provided with the
distribution.
3. All advertising materials mentioning features or use of this
software must display the following acknowledgment:
"This product includes software developed by the OpenSSL Project
for use in the OpenSSL Toolkit. (http://www.openssl.org/)"
4. The names "OpenSSL Toolkit" and "OpenSSL Project" must not be used to
endorse or promote products derived from this software without
prior written permission. For written permission, please contact
[email protected].
5. Products derived from this software may not be called "OpenSSL"
nor may "OpenSSL" appear in their names without prior written
permission of the OpenSSL Project.
6. Redistributions of any form whatsoever must retain the following
acknowledgment:
"This product includes software developed by the OpenSSL Project
for use in the OpenSSL Toolkit (http://www.openssl.org/)"
THIS SOFTWARE IS PROVIDED BY THE OpenSSL PROJECT ``AS IS'' AND ANY
EXPRESSED OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR
PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE OpenSSL PROJECT OR
ITS CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL,
SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT
NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES;
LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION)
HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT,
STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE)
ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED
OF THE POSSIBILITY OF SUCH DAMAGE.
====================================================================
This product includes cryptographic software written by Eric Young
([email protected]). This product includes software written by Tim
Hudson ([email protected]).
Original SSLeay License
-----------------------
Copyright (C) 1995-1998 Eric Young ([email protected])
All rights reserved.
This package is an SSL implementation written
by Eric Young ([email protected]).
The implementation was written so as to conform with Netscapes SSL.
This library is free for commercial and non-commercial use as long as
the following conditions are aheared to. The following conditions
apply to all code found in this distribution, be it the RC4, RSA,
lhash, DES, etc., code; not just the SSL code. The SSL documentation
included with this distribution is covered by the same copyright terms
except that the holder is Tim Hudson ([email protected]).
Copyright remains Eric Young's, and as such any Copyright notices in
the code are not to be removed.
If this package is used in a product, Eric Young should be given attribution
as the author of the parts of the library used.
This can be in the form of a textual message at program startup or
in documentation (online or textual) provided with the package.
Redistribution and use in source and binary forms, with or without
modification, are permitted provided that the following conditions
are met:
1. Redistributions of source code must retain the copyright
notice, this list of conditions and the following disclaimer.
2. Redistributions in binary form must reproduce the above copyright
notice, this list of conditions and the following disclaimer in the
documentation and/or other materials provided with the distribution.
3. All advertising materials mentioning features or use of this software
must display the following acknowledgement:
"This product includes cryptographic software written by
Eric Young ([email protected])"
The word 'cryptographic' can be left out if the rouines from the library
being used are not cryptographic related :-).
4. If you include any Windows specific code (or a derivative thereof) from
the apps directory (application code) you must include an acknowledgement:
"This product includes software written by Tim Hudson ([email protected])"
THIS SOFTWARE IS PROVIDED BY ERIC YOUNG ``AS IS'' AND
ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE
ARE DISCLAIMED. IN NO EVENT SHALL THE AUTHOR OR CONTRIBUTORS BE LIABLE
FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS
OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION)
HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT
LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY
OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF
SUCH DAMAGE.
The licence and distribution terms for any publically available version or
derivative of this code cannot be changed. i.e. this code cannot simply be
copied and put under another distribution licence
[including the GNU Public Licence.]