jonniedie / Componentarrays.jl
Programming Languages
Projects that are alternatives of or similar to Componentarrays.jl
ComponentArrays.jl
| Documentation | Build Status |
|---|---|
 
|
|
The main export of this package is the ComponentArray type. "Components" of ComponentArrays
are really just array blocks that can be accessed through a named index. This will create a new ComponentArray whose data is a view into the original,
allowing for standalone models to be composed together by simple function composition. In
essence, ComponentArrays allow you to do the things you would usually need a modeling
language for, but without actually needing a modeling language. The main targets are for use
in DifferentialEquations.jl and
Optim.jl, but anything that requires
flat vectors is fair game.
General use
The easiest way to construct 1-dimensional ComponentArrays (aliased as ComponentVector) is as if they were NamedTuples. In fact, a good way to think about them is as arbitrarily nested, mutable NamedTuples that can be passed through a solver.
julia> c = (a=2, b=[1, 2]);
julia> x = ComponentArray(a=5, b=[(a=20., b=0), (a=33., b=0), (a=44., b=3)], c=c)
ComponentVector{Float64}(a = 5.0, b = [(a = 20.0, b = 0.0), (a = 33.0, b = 0.0), (a = 44.0, b = 3.0)], c = (a = 2.0, b = [1.0, 2.0]))
julia> x.c.a = 400; x
ComponentVector{Float64}(a = 5.0, b = [(a = 20.0, b = 0.0), (a = 33.0, b = 0.0), (a = 44.0, b = 3.0)], c = (a = 400.0, b = [1.0, 2.0]))
julia> x[8]
400.0
julia> collect(x)
10-element Array{Float64,1}:
5.0
20.0
0.0
33.0
0.0
44.0
3.0
400.0
1.0
2.0
julia> typeof(similar(x, Int32)) === typeof(ComponentVector{Int32}(a=5, b=[(a=20., b=0), (a=33., b=0), (a=44., b=3)], c=c))
true
Higher dimensional ComponentArrays can be created too, but it's a little messy at the moment. The nice thing for modeling is that dimension expansion through broadcasted operations can create higher-dimensional ComponentArrays automatically, so Jacobian cache arrays that are created internally with false .* x .* x' will be two-dimensional ComponentArrays (aliased as ComponentMatrix) with proper axes. Check out the ODE with Jacobian example in the examples folder to see how this looks in practice.
julia> x = ComponentArray(a=1, b=[2, 1, 4.0], c=c)
ComponentVector{Float64}(a = 1.0, b = [2.0, 1.0, 4.0], c = (a = 2.0, b = [1.0, 2.0]))
julia> x2 = x .* x'
7×7 ComponentMatrix{Float64} with axes Axis(a = 1, b = 2:4, c = ViewAxis(5:7, Axis(a = 1, b = 2:3))) × Axis(a = 1, b = 2:4, c = ViewAxis(5:7, Axis(a = 1, b = 2:3)))
1.0 2.0 1.0 4.0 2.0 1.0 2.0
2.0 4.0 2.0 8.0 4.0 2.0 4.0
1.0 2.0 1.0 4.0 2.0 1.0 2.0
4.0 8.0 4.0 16.0 8.0 4.0 8.0
2.0 4.0 2.0 8.0 4.0 2.0 4.0
1.0 2.0 1.0 4.0 2.0 1.0 2.0
2.0 4.0 2.0 8.0 4.0 2.0 4.0
julia> x2[:c,:c]
3×3 ComponentMatrix{Float64,SubArray...} with axes Axis(a = 1, b = 2:3) × Axis(a = 1, b = 2:3)
4.0 2.0 4.0
2.0 1.0 2.0
4.0 2.0 4.0
julia> x2[:a,:a]
1.0
julia> x2[:a,:c]
ComponentVector{Float64,SubArray...}(a = 2.0, b = [1.0, 2.0])
julia> x2[:b,:c]
3×3 ComponentMatrix{Float64,SubArray...} with axes FlatAxis() × Axis(a = 1, b = 2:3)
4.0 2.0 4.0
2.0 1.0 2.0
8.0 4.0 8.0
Examples
Differential equation example
This example uses @unpack from Parameters.jl
for nice syntax. Example taken from:
https://github.com/JuliaDiffEq/ModelingToolkit.jl/issues/36#issuecomment-536221300
using ComponentArrays
using DifferentialEquations
using Parameters: @unpack
tspan = (0.0, 20.0)
## Lorenz system
function lorenz!(D, u, p, t; f=0.0)
@unpack σ, ρ, β = p
@unpack x, y, z = u
D.x = σ*(y - x)
D.y = x*(ρ - z) - y - f
D.z = x*y - β*z
return nothing
end
lorenz_p = (σ=10.0, ρ=28.0, β=8/3)
lorenz_ic = ComponentArray(x=0.0, y=0.0, z=0.0)
lorenz_prob = ODEProblem(lorenz!, lorenz_ic, tspan, lorenz_p)
## Lotka-Volterra system
function lotka!(D, u, p, t; f=0.0)
@unpack α, β, γ, δ = p
@unpack x, y = u
D.x = α*x - β*x*y + f
D.y = -γ*y + δ*x*y
return nothing
end
lotka_p = (α=2/3, β=4/3, γ=1.0, δ=1.0)
lotka_ic = ComponentArray(x=1.0, y=1.0)
lotka_prob = ODEProblem(lotka!, lotka_ic, tspan, lotka_p)
## Composed Lorenz and Lotka-Volterra system
function composed!(D, u, p, t)
c = p.c #coupling parameter
@unpack lorenz, lotka = u
lorenz!(D.lorenz, lorenz, p.lorenz, t, f=c*lotka.x)
lotka!(D.lotka, lotka, p.lotka, t, f=c*lorenz.x)
return nothing
end
comp_p = (lorenz=lorenz_p, lotka=lotka_p, c=0.01)
comp_ic = ComponentArray(lorenz=lorenz_ic, lotka=lotka_ic)
comp_prob = ODEProblem(composed!, comp_ic, tspan, comp_p)
## Solve problem
# We can solve the composed system...
comp_sol = solve(comp_prob)
# ...or we can unit test one of the component systems
lotka_sol = solve(lotka_prob)
Notice how cleanly the composed! function can pass variables from one function to another with no array index juggling in sight. This is especially useful for large models as it becomes harder to keep track top-level model array position when adding new or deleting old components from the model. We could go further and compose composed! with other components ad (practically) infinitum with no mental bookkeeping.
The main benefit, however, is now our differential equations are unit testable. Both lorenz and lotka can be run as their own ODEProblem with f set to zero to see the unforced response.
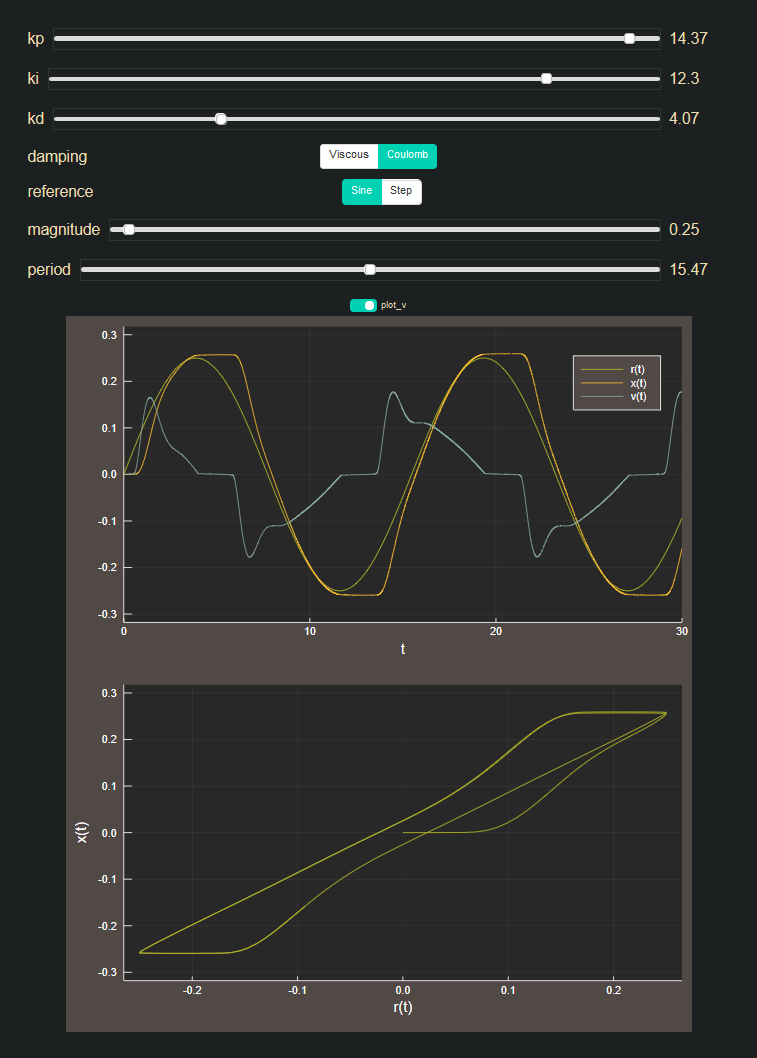
Control of a sliding block
In this example, we'll build a model of a block sliding on a surface and use ComponentArrays to easily switch between coulomb and equivalent viscous damping models. The block is controlled by pushing and pulling a spring attached to it and we will use feedback through a PID controller to try to track a reference signal. For simplification, we are using the velocity of the block directly for the derivative term, rather than taking a filtered derivative of the error signal. We are also setting a deadzone on the friction force with exponential decay to zero velocity to get rid of simulation chatter during the static friction regime.
using ComponentArrays
using DifferentialEquations
using Interact: @manipulate
using Parameters: @unpack
using Plots
## Setup
const g = 9.80665
maybe_apply(f::Function, x, p, t) = f(x, p, t)
maybe_apply(f, x, p, t) = f
# Allows functions of form f(x,p,t) to be applied and passed in as inputs
function simulator(func; kwargs...)
simfun(dx, x, p, t) = func(dx, x, p, t; map(f->maybe_apply(f, x, p, t), (;kwargs...))...)
simfun(x, p, t) = func(x, p, t; map(f->maybe_apply(f, x, p, t), (;kwargs...))...)
return simfun
end
softsign(x) = tanh(1e3x)
## Dynamics update functions
# Sliding block with viscous friction
function viscous_block!(D, vars, p, t; u=0.0)
@unpack m, c, k = p
@unpack v, x = vars
D.x = v
D.v = (-c*v + k*(u-x))/m
return x
end
# Sliding block with coulomb friction
function coulomb_block!(D, vars, p, t; u=0.0)
@unpack m, μ, k = p
@unpack v, x = vars
D.x = v
a = -μ*g*softsign(v) + k*(u-x)/m
D.v = abs(a)<1e-3 && abs(v)<1e-3 ? -10v : a
return x
end
function PID_controller!(D, vars, p, t; err=0.0, v=0.0)
@unpack kp, ki, kd = p
@unpack x = vars
D.x = err
return ki*x + kp*err + kd*v
end
function feedback_sys!(D, components, p, t; ref=0.0)
@unpack ctrl, plant = components
u = p.ctrl.fun(D.ctrl, ctrl, p.ctrl.params, t; err=ref-plant.x, v=-plant.v)
return p.plant.fun(D.plant, plant, p.plant.params, t; u=u)
end
step_input(;time=1.0, mag=1.0) = (x,p,t) -> t>time ? mag : 0
sine_input(;mag=1.0, period=10.0) = (x,p,t) -> mag*sin(t*2π/period)
## Interactive GUI for switching out plant models and varying PID gains
@manipulate for kp in 0:0.01:15,
ki in 0:0.01:15,
kd in 0:0.01:15,
damping in Dict(
"Coulomb" => coulomb_block!,
"Viscous" => viscous_block!,
),
reference in Dict(
"Sine" => sine_input,
"Step" => step_input,
),
magnitude in 0:0.01:10, # pop-pop!
period in 1:0.01:30,
plot_v in false
# Inputs
tspan = (0.0, 30.0)
ctrl_fun = PID_controller!
# plant_fun = coulomb_block!
ref = if reference==sine_input
reference(period=period, mag=magnitude)
else
reference(mag=magnitude)
end
m = 50.0
μ = 0.1
ω = 2π/period
c = 4*μ*m*g/(π*ω*magnitude) # Viscous equivalent damping
k = 50.0
plant_p = (m=m, μ=μ, c=c, k=k)
ctrl_p = (kp=kp, ki=ki, kd=kd)
plant_ic = (v=0, x=0)
ctrl_ic = (;x=0)
# Set up and solve
sys_p = (
ctrl = (
params = ctrl_p,
fun = ctrl_fun,
),
plant = (
params = plant_p,
fun = damping,
),
)
sys_ic = ComponentArray(ctrl=ctrl_ic, plant=plant_ic)
sys_fun = ODEFunction(simulator(feedback_sys!, ref=ref), syms=[:u, :v, :x])
sys_prob = ODEProblem(sys_fun, sys_ic, tspan, sys_p)
sol = solve(sys_prob, Tsit5())
# Plot
t = tspan[1]:0.1:tspan[2]
lims = magnitude*[-1, 1]
plotvars = plot_v ? [3, 2] : [3]
strip = plot(t, ref.(0, 0, t), ylim=1.2lims, label="r(t)")
plot!(strip, sol, vars=plotvars)
phase = plot(ref.(0, 0, t), map(x->x.plant.x, sol(t).u),
xlim=lims,
ylim=1.2lims,
legend=false,
xlabel="r(t)",
ylabel="x(t)",
)
plot(strip, phase, layout=(2, 1), size=(700, 800))
end