zhwhong / Lidc_nodule_detection
Projects that are alternatives of or similar to Lidc nodule detection
Introduction
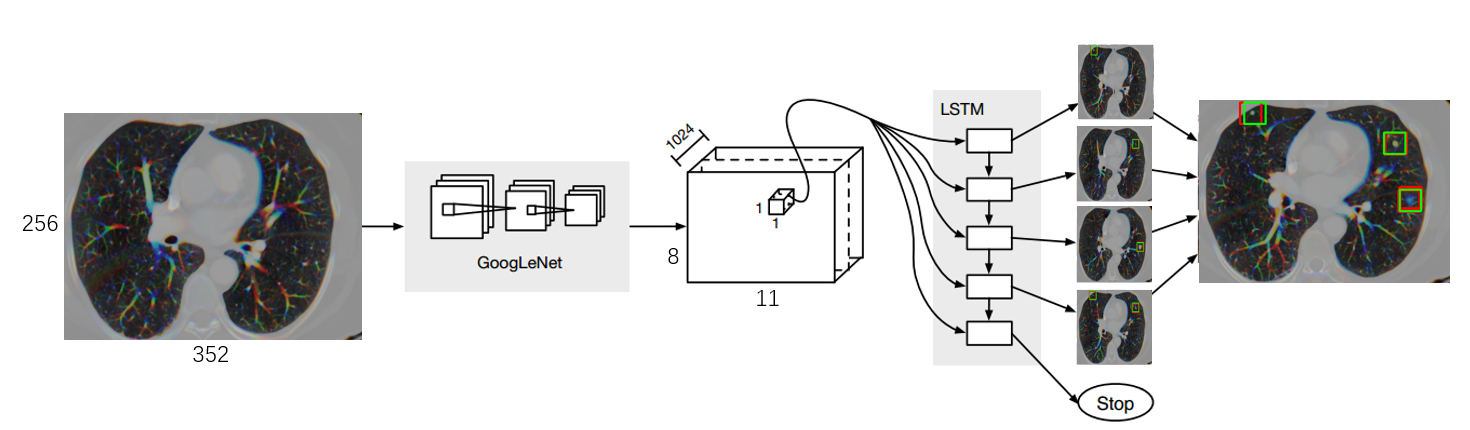
This is a simple framework for training neural networks to detect nodules in CT images. Training requires a json file (e.g. here) containing a list of CT images and the bounding boxes in each image. The model combines both CNN model and LSTM unit. The algorithm here is mainly refered to paper End-to-end people detection in crowded scenes.
The deep learning framewoek is based on TensorFlow(version 1.0.0) and some coding ideas are forked from the TensorBox project. Here I show heartfelt gratefulness. About nodule classfication method based on CNN transfer learning, you can refer to this paper.
Dicom and XML parsing
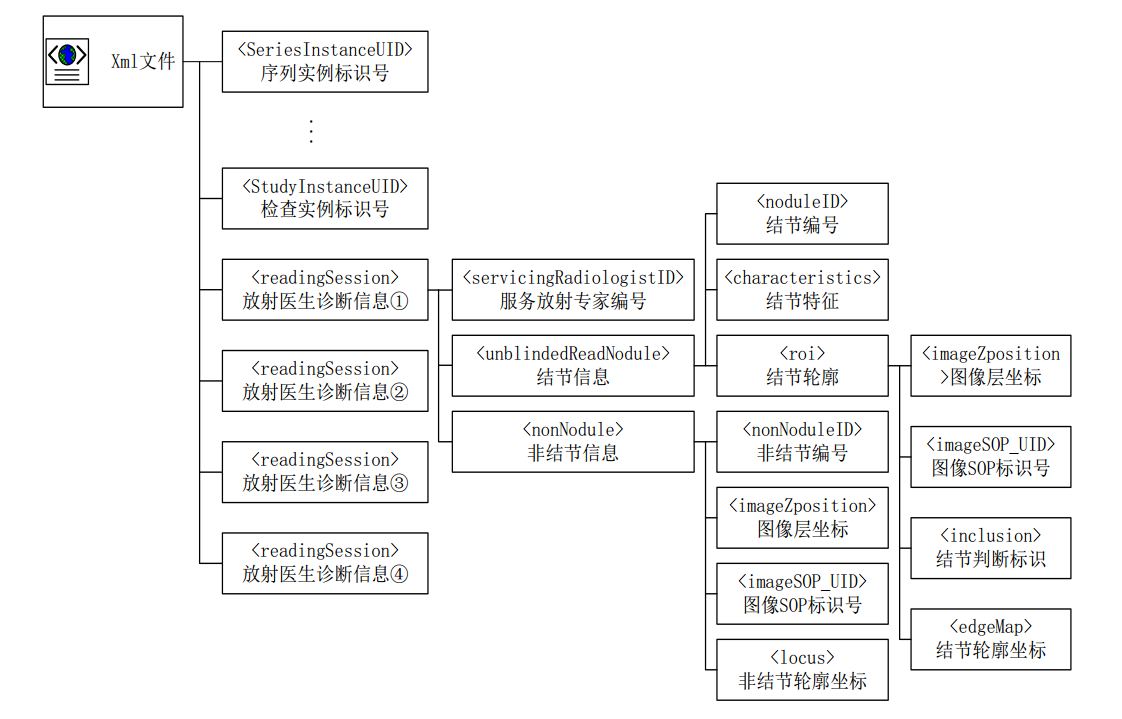
Parsing the lidc XML annotation and dicom files,see pylung and this blog.
>>> import dicom
>>> f = dicom.read_file('000001.dcm')
>>> print f
(0008, 0005) Specific Character Set CS: 'ISO_IR 100'
(0008, 0008) Image Type CS: ['ORIGINAL', 'PRIMARY', 'AXIAL']
(0008, 0016) SOP Class UID UI: CT Image Storage
(0008, 0018) SOP Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.143451261327128179989900675595
(0008, 0020) Study Date DA: '20000101'
(0008, 0021) Series Date DA: '20000101'
(0008, 0022) Acquisition Date DA: '20000101'
(0008, 0023) Content Date DA: '20000101'
(0008, 0024) Overlay Date DA: '20000101'
(0008, 0025) Curve Date DA: '20000101'
(0008, 002a) Acquisition DateTime DT: '20000101'
(0008, 0030) Study Time TM: ''
(0008, 0032) Acquisition Time TM: ''
(0008, 0033) Content Time TM: ''
(0008, 0050) Accession Number SH: '2819497684894126'
(0008, 0060) Modality CS: 'CT'
(0008, 0070) Manufacturer LO: 'GE MEDICAL SYSTEMS'
(0008, 0090) Referring Physician Name PN: ''
(0008, 1090) Manufacturer Model Name LO: 'LightSpeed Plus'
(0008, 1155) Referenced SOP Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.675906998158803995297223798692
(0010, 0010) Patient Name PN: ''
(0010, 0020) Patient ID LO: 'LIDC-IDRI-0001'
(0010, 0030) Patient Birth Date DA: ''
(0010, 0040) Patient Sex CS: ''
(0010, 1010) Patient Age AS: ''
(0010, 21d0) Last Menstrual Date DA: '20000101'
(0012, 0062) Patient Identity Removed CS: 'YES'
(0012, 0063) De-identification Method LO: 'DCM:113100/113105/113107/113108/113109/113111'
(0013, 0010) Private Creator LO: 'CTP'
(0013, 1010) Private tag data LO: 'LIDC-IDRI'
(0013, 1013) Private tag data LO: '62796001'
(0018, 0010) Contrast/Bolus Agent LO: 'IV'
(0018, 0015) Body Part Examined CS: 'CHEST'
(0018, 0022) Scan Options CS: 'HELICAL MODE'
(0018, 0050) Slice Thickness DS: '2.500000'
(0018, 0060) KVP DS: '120'
(0018, 0090) Data Collection Diameter DS: '500.000000'
(0018, 1020) Software Version(s) LO: 'LightSpeedApps2.4.2_H2.4M5'
(0018, 1100) Reconstruction Diameter DS: '360.000000'
(0018, 1110) Distance Source to Detector DS: '949.075012'
(0018, 1111) Distance Source to Patient DS: '541.000000'
(0018, 1120) Gantry/Detector Tilt DS: '0.000000'
(0018, 1130) Table Height DS: '144.399994'
(0018, 1140) Rotation Direction CS: 'CW'
(0018, 1150) Exposure Time IS: '570'
(0018, 1151) X-Ray Tube Current IS: '400'
(0018, 1152) Exposure IS: '4684'
(0018, 1160) Filter Type SH: 'BODY FILTER'
(0018, 1170) Generator Power IS: '48000'
(0018, 1190) Focal Spot(s) DS: '1.200000'
(0018, 1210) Convolution Kernel SH: 'STANDARD'
(0018, 5100) Patient Position CS: 'FFS'
(0020, 000d) Study Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.298806137288633453246975630178
(0020, 000e) Series Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.179049373636438705059720603192
(0020, 0010) Study ID SH: ''
(0020, 0011) Series Number IS: '3000566'
(0020, 0013) Instance Number IS: '80'
(0020, 0032) Image Position (Patient) DS: ['-166.000000', '-171.699997', '-207.500000']
(0020, 0037) Image Orientation (Patient) DS: ['1.000000', '0.000000', '0.000000', '0.000000', '1.000000', '0.000000']
(0020, 0052) Frame of Reference UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.229925374658226729607867499499
(0020, 1040) Position Reference Indicator LO: 'SN'
(0020, 1041) Slice Location DS: '-207.500000'
(0028, 0002) Samples per Pixel US: 1
(0028, 0004) Photometric Interpretation CS: 'MONOCHROME2'
(0028, 0010) Rows US: 512
(0028, 0011) Columns US: 512
(0028, 0030) Pixel Spacing DS: ['0.703125', '0.703125']
(0028, 0100) Bits Allocated US: 16
(0028, 0101) Bits Stored US: 16
(0028, 0102) High Bit US: 15
(0028, 0103) Pixel Representation US: 1
(0028, 0120) Pixel Padding Value US: 63536
(0028, 0303) Longitudinal Temporal Information M CS: 'MODIFIED'
(0028, 1050) Window Center DS: '-600'
(0028, 1051) Window Width DS: '1600'
(0028, 1052) Rescale Intercept DS: '-1024'
(0028, 1053) Rescale Slope DS: '1'
(0038, 0020) Admitting Date DA: '20000101'
(0040, 0002) Scheduled Procedure Step Start Date DA: '20000101'
(0040, 0004) Scheduled Procedure Step End Date DA: '20000101'
(0040, 0244) Performed Procedure Step Start Date DA: '20000101'
(0040, 2016) Placer Order Number / Imaging Servi LO: ''
(0040, 2017) Filler Order Number / Imaging Servi LO: ''
(0040, a075) Verifying Observer Name PN: 'Removed by CTP'
(0040, a123) Person Name PN: 'Removed by CTP'
(0040, a124) UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.335419887712224178340067932923
(0070, 0084) Content Creator's Name PN: ''
(0088, 0140) Storage Media File-set UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.211790042620307056609660772296
(7fe0, 0010) Pixel Data OW: Array of 524288 bytes
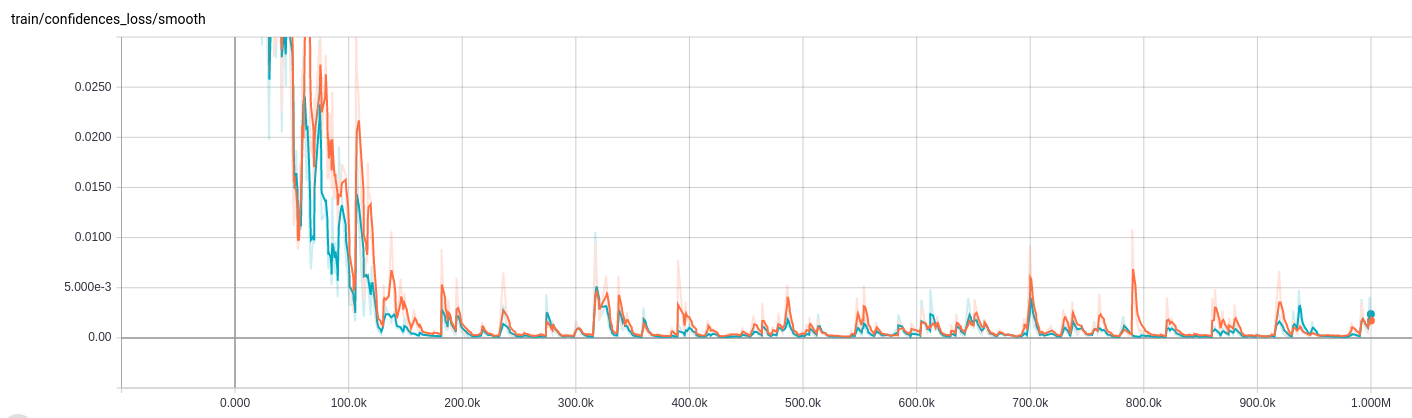
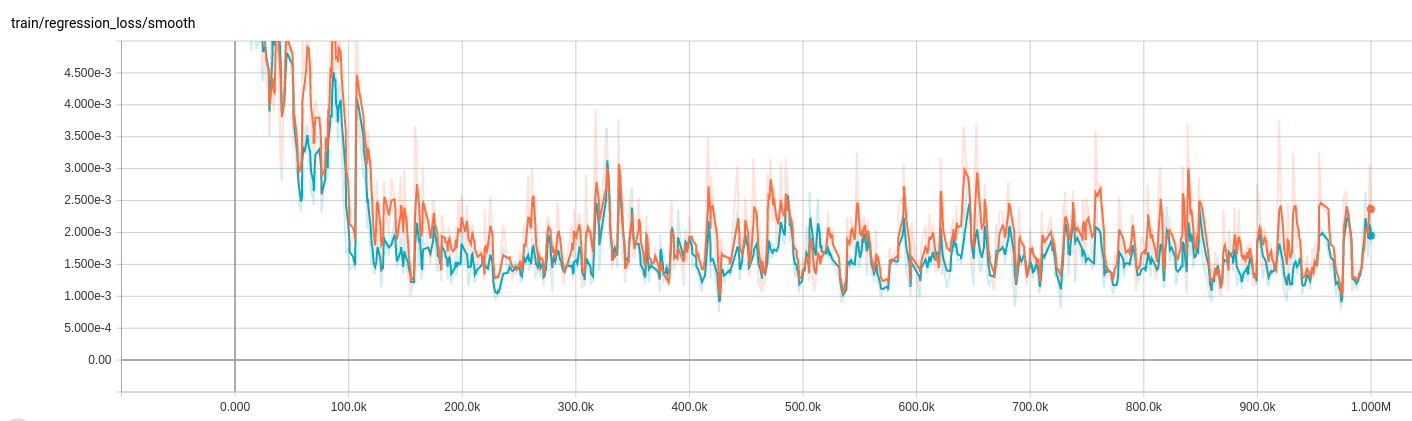
Training
First, install TensorFlow from source or pip (NB: source installs currently break threading on 0.11). Then run the training script CNN_LSTM/run.sh.Note that running on your own dataset should require modifying the CNN_LSTM/hypes/\*.json file.
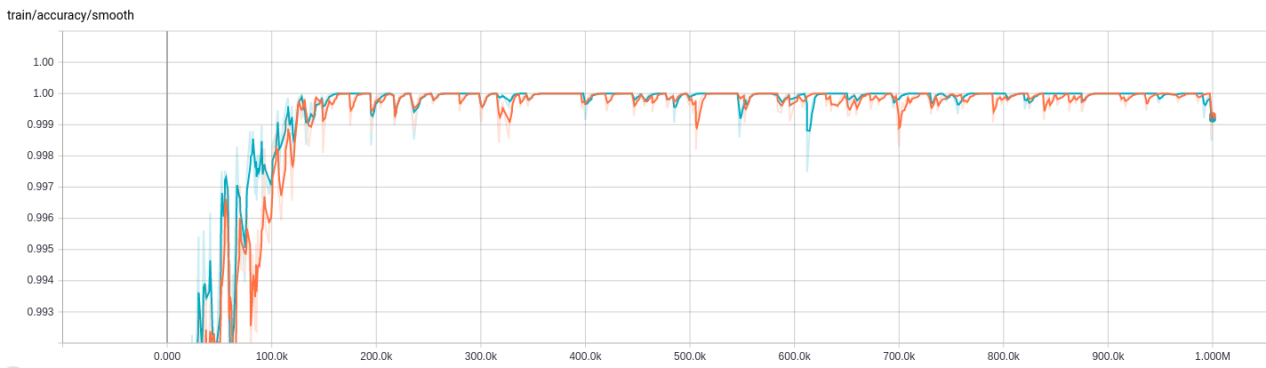
Evaluation
There are two options for evaluation, an ipython notebook and a python script.
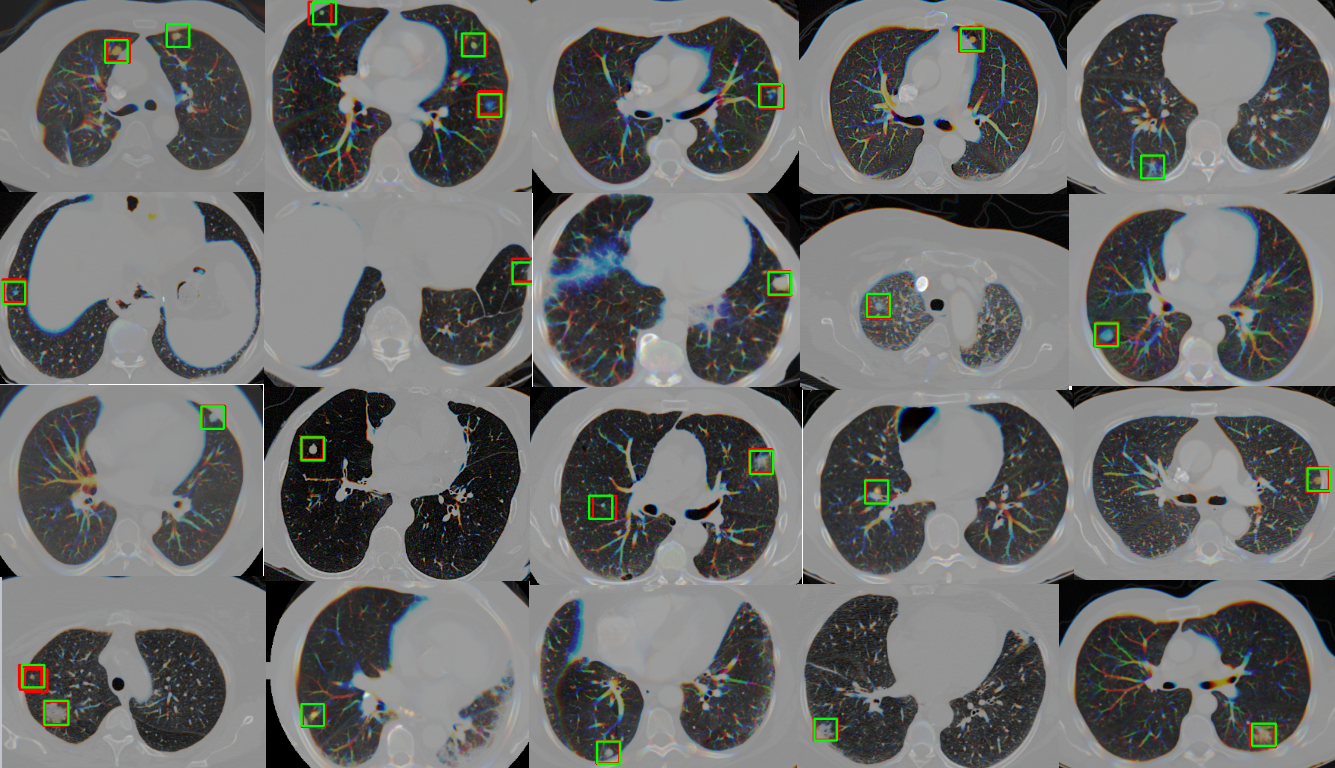
- some test results
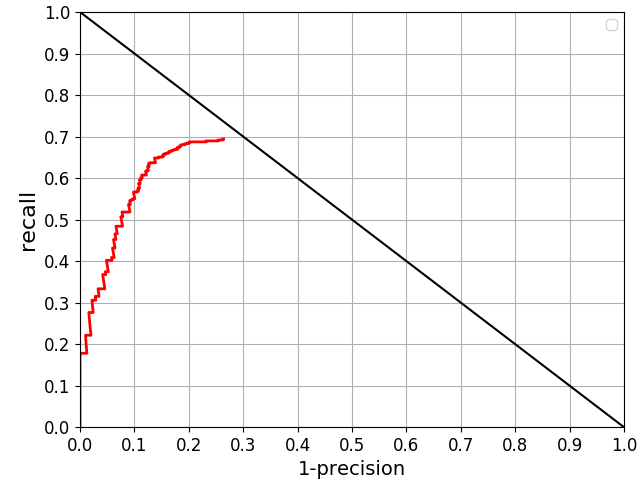
- precision-recall curve
Other
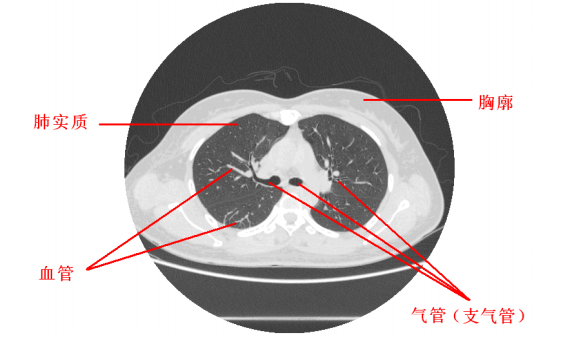
- Anatomy map
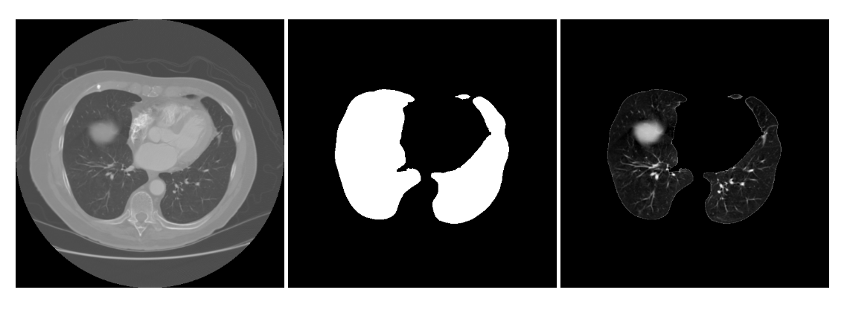
- Parenchyma extraction
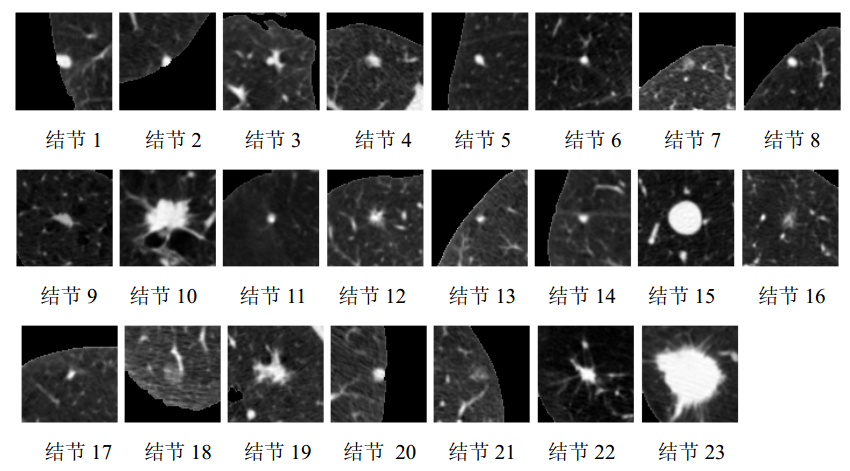
- Pulmonary nodules form
Reference
- [Dataset] The Lung Image Database Consortium image collection(LIDC-IDRI)
- [Paper] End-to-end people detection in crowded scenes
- [Paper] Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning
- [Paper] An overview of classification algorithms for imbalanced datasets
- [Blog] LIDC-IDRI肺结节公开数据集Dicom和XML标注详解
- [Blog] 机器学习之分类性能度量指标 : ROC曲线、AUC值、正确率、召回率
- [简书] LIDC-IDRI肺结节Dicom数据集解析与总结
- [简书] LIDC-IDRI肺结节公开数据集Dicom和XML标注详解
- [简书] 医疗CT影像肺结节检测参考项目(附论文)
- [简书] 如何应用Python处理医学影像学中的DICOM信息
- [简书] CT图像肺结节识别算法调研 — CNN篇
- [简书] 吕乐:面向医学图像计算的深度学习与卷积神经网络(转)