joiningdata / Lollipops
Programming Languages
Projects that are alternatives of or similar to Lollipops
lollipops
Please cite this work as
- Jay JJ, Brouwer C (2016) Lollipops in the Clinic: Information Dense Mutation Plots for Precision Medicine. PLoS ONE 11(8): e0160519. doi: 10.1371/journal.pone.0160519.
A simple 'lollipop' mutation diagram generator that tries to make things simple and easy by automating as much as possible. It uses the UniProt REST API and/or Pfam API to automate translation of Gene Symbols and lookup domain/motif features for display. If variant changes are provided, it will also annotate them to the diagram using the "lollipops" markers that give the tool it's name.
Example
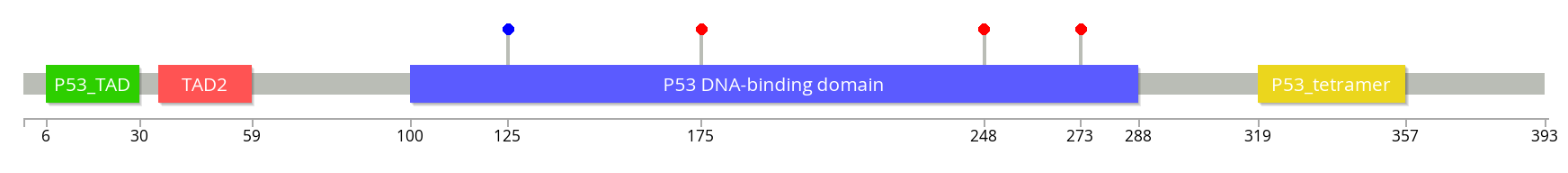
Basic usage is just the gene symbol (ex: TP53) and a list of
mutations (ex: R273C R175H T125 R248Q)
./lollipops TP53 R273C R175H T125 R248Q
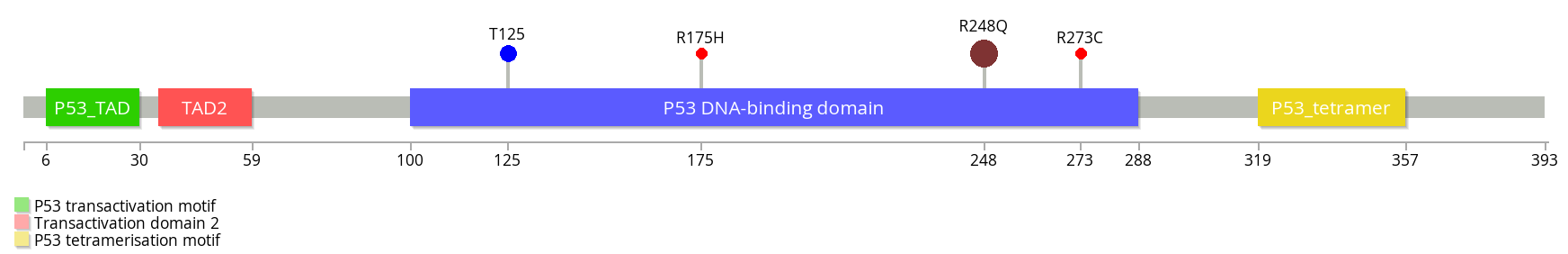
More advanced usage allows for per-mutation color (e.g. sample type) and size specification (e.g. denoting number of samples), along with text labels, a legend for abbreviated domains, and more:
./lollipops -legend -labels TP53 R248Q#[email protected] R273C R175H [email protected]
Usage
Usage: lollipops [options] {-U UNIPROT_ID | GENE_SYMBOL} [PROTEIN CHANGES ...]
Where GENE_SYMBOL is the official human HGNC gene symbol. This will use the
official API to lookup the UNIPROT_ID. To skip the lookup or use other species,
specify the UniProt ID with -U (e.g. -U P04637 for TP53)
Protein changes
Currently only point mutations are supported, and may be specified as:
<AMINO><CODON><AMINO><#COLOR><@COUNT>
Only CODON is required, and AMINO tags are not parsed.
Synonymous mutations are denoted if the first AMINO tag matches the second AMINO tag, or if the second tag is not present. Otherwise the non-synonymous mutation color is used. The COLOR tag will override using the #RRGGBB style provided. The COUNT tag can be used to scale the lollipop marker size so that the area is exponentially proportional to the count indicated. Examples:
R273C -- non-synonymous mutation at codon 273
[email protected] -- synonymous mutation at codon 125 with "5x" marker sizing
R248Q#00ff00 -- green lollipop at codon 248
R248Q#[email protected] -- green lollipop at codon 248 with "131x" marker sizing
N.B. Color must come before count in tags.
Diagram generation options
-legend draw a legend for colored regions
-syn-color="#0000ff" color to use for synonymous mutation markers
-mut-color="#ff0000" color to use for non-synonymous mutation markers
-hide-axis do not draw the amino position x-axis
-show-disordered draw disordered regions on the backbone
-show-motifs draw simple motif regions
-labels draw label text above lollipop markers
-no-patterns use solid fill instead of patterns (SVG only)
Output options
-o=filename.png set output filename (.png or .svg supported)
-w=700 set diagram pixel width (default = automatic fit)
-dpi=300 set DPI (PNG output only)
Alternative input sources:
-uniprot use UniprotKB as an alternative to Pfam for
fetching domain/motif information
-l=filename.json use local file instead of Pfam API for graphic data
see: http://pfam.xfam.org/help#tabview=tab9
Installation
Head over to the Releases to download the latest version for your system in a simple command-line executable.
If you already have Go installed and want the bleeding edge, just
go get -u github.com/joiningdata/lollipops to download the latest version.
Embedding
As of v0.97, lollipops is now easy to embed in other Go applications. The following code prints an SVG for TP53 and some mutations to standard output:
package main
import (
"os"
"github.com/joiningdata/lollipops/data"
"github.com/joiningdata/lollipops/drawing"
)
func main() {
uniprot_id := "P04637"
mutations := []string{"R273C", "R175H", "[email protected]"}
p53_domains, err := data.GetGraphicData(uniprot_id)
if err != nil {
panic(err)
}
drawing.DrawSVG(os.Stdout, mutations, p53_domains)
}
CONTRIBUTING
Please submit your bugs and features requests via the Issues tab. Be sure to search closed issues before submitting a new one in case the issue has been previously discussed. Pull Requests are welcome, but please create an issue beforehand to discuss significant changes.
Code contributions are expected to be properly formatted with go fmt, and generally adhere to the standard Golang review guidelines.
LICENSE
Lollipops is released under the GPL license. If your institution requires additional licensing options please contact the author.