PaddlePaddle / Paddlehelix
Licence: apache-2.0
Bio-Computing Platform featuring Large-Scale Representation Learning and Multi-Task Deep Learning “螺旋桨”生物计算工具集
Stars: ✭ 213
Projects that are alternatives of or similar to Paddlehelix
Codesearchnet
Datasets, tools, and benchmarks for representation learning of code.
Stars: ✭ 1,378 (+546.95%)
Mutual labels: jupyter-notebook, representation-learning
Cutout Random Erasing
Cutout / Random Erasing implementation, especially for ImageDataGenerator in Keras
Stars: ✭ 142 (-33.33%)
Mutual labels: jupyter-notebook, deeplearning
Sigver wiwd
Learned representation for Offline Handwritten Signature Verification. Models and code to extract features from signature images.
Stars: ✭ 112 (-47.42%)
Mutual labels: jupyter-notebook, representation-learning
Text Classification
Text Classification through CNN, RNN & HAN using Keras
Stars: ✭ 216 (+1.41%)
Mutual labels: jupyter-notebook, deeplearning
Fixy
Amacımız Türkçe NLP literatüründeki birçok farklı sorunu bir arada çözebilen, eşsiz yaklaşımlar öne süren ve literatürdeki çalışmaların eksiklerini gideren open source bir yazım destekleyicisi/denetleyicisi oluşturmak. Kullanıcıların yazdıkları metinlerdeki yazım yanlışlarını derin öğrenme yaklaşımıyla çözüp aynı zamanda metinlerde anlamsal analizi de gerçekleştirerek bu bağlamda ortaya çıkan yanlışları da fark edip düzeltebilmek.
Stars: ✭ 165 (-22.54%)
Mutual labels: jupyter-notebook, deeplearning
Spectralnormalizationkeras
Spectral Normalization for Keras Dense and Convolution Layers
Stars: ✭ 100 (-53.05%)
Mutual labels: jupyter-notebook, deeplearning
All4nlp
All For NLP, especially Chinese.
Stars: ✭ 141 (-33.8%)
Mutual labels: jupyter-notebook, deeplearning
Machine learning code
机器学习与深度学习算法示例
Stars: ✭ 88 (-58.69%)
Mutual labels: jupyter-notebook, deeplearning
Motion Sense
MotionSense Dataset for Human Activity and Attribute Recognition ( time-series data generated by smartphone's sensors: accelerometer and gyroscope)
Stars: ✭ 159 (-25.35%)
Mutual labels: jupyter-notebook, deeplearning
Anomaly detection tuto
Anomaly detection tutorial on univariate time series with an auto-encoder
Stars: ✭ 144 (-32.39%)
Mutual labels: jupyter-notebook, deeplearning
Sert
Semantic Entity Retrieval Toolkit
Stars: ✭ 100 (-53.05%)
Mutual labels: deeplearning, representation-learning
Release
Deep Reinforcement Learning for de-novo Drug Design
Stars: ✭ 201 (-5.63%)
Mutual labels: jupyter-notebook, deeplearning
Ngsim env
Learning human driver models from NGSIM data with imitation learning.
Stars: ✭ 96 (-54.93%)
Mutual labels: jupyter-notebook, deeplearning
Pytorch Byol
PyTorch implementation of Bootstrap Your Own Latent: A New Approach to Self-Supervised Learning
Stars: ✭ 213 (+0%)
Mutual labels: jupyter-notebook, representation-learning
Classification Of Hyperspectral Image
Classification of the Hyperspectral Image Indian Pines with Convolutional Neural Network
Stars: ✭ 93 (-56.34%)
Mutual labels: jupyter-notebook, deeplearning
Seq2seq tutorial
Code For Medium Article "How To Create Data Products That Are Magical Using Sequence-to-Sequence Models"
Stars: ✭ 132 (-38.03%)
Mutual labels: jupyter-notebook, deeplearning
Mit Deep Learning
Tutorials, assignments, and competitions for MIT Deep Learning related courses.
Stars: ✭ 8,912 (+4084.04%)
Mutual labels: jupyter-notebook, deeplearning
Novel Deep Learning Model For Traffic Sign Detection Using Capsule Networks
capsule networks that achieves outstanding performance on the German traffic sign dataset
Stars: ✭ 88 (-58.69%)
Mutual labels: jupyter-notebook, deeplearning
Deep Learning With Tensorflow Book
深度学习入门开源书,基于TensorFlow 2.0案例实战。Open source Deep Learning book, based on TensorFlow 2.0 framework.
Stars: ✭ 12,105 (+5583.1%)
Mutual labels: jupyter-notebook, deeplearning
Simclr
SimCLRv2 - Big Self-Supervised Models are Strong Semi-Supervised Learners
Stars: ✭ 2,720 (+1177%)
Mutual labels: jupyter-notebook, representation-learning
English | 简体中文
PaddleHelix is a machine-learning-based bio-computing framework aiming at facilitating the development of the following areas:
- Vaccine design
- Drug discovery
- Precision medicine
Features
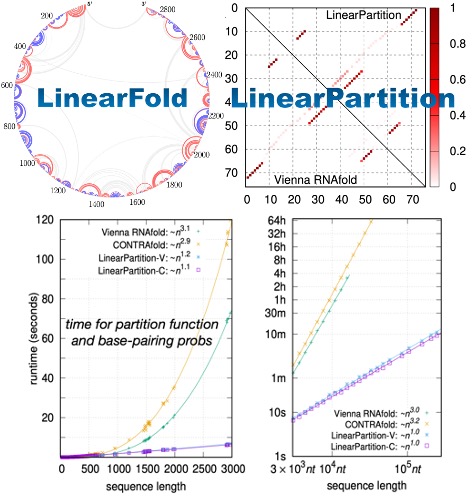
- High Efficency: We provide LinearRNA, a highly efficient toolkit for RNA structure prediction and analysis. LinearFold & LinearPartition achieve O(n) complexity in RNA-folding prediction, which is hundreds of times faster than traditional folding techniques.
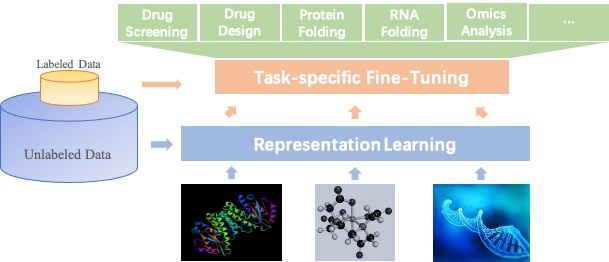
- Large-scale Representation Learning and Transfer Learning: Self-supervised learning for molecule representations offers prospects of a breakthrough in tasks with limited annotation, including drug profiling, drug-target interaction, protein-protein interaction, RNA-RNA interaction, protein folding, RNA folding, and molecule design. PaddleHelix implements a variety of representation learning algorithms and state-of-the-art large-scale pre-trained models to help developers to start from "the shoulders of giants" quickly.
- Easy-to-use APIs: PaddleHelix provides frequently used structures and pre-trained models. You can easily use those components to build up your models and systems.
Installation
The installation prerequisites and guide can be found here.
Documentation
Tutorials
- We provide abundant tutorials to help you navigate the repository and start quickly.
- PaddleHelix is based on PaddlePaddle, a high-performance Parallelized Deep Learning Platform.
Examples
- Representation Learning - Compounds
- Representation Learning - Proteins
- Drug-Target Interaction
- LinearRNA
The API reference
- Detailed API reference of PaddleHelix can be found here.
Guide for developers
- If you need help in modifying the source code of PaddleHelix, please see our Guide for developers.
Note that the project description data, including the texts, logos, images, and/or trademarks,
for each open source project belongs to its rightful owner.
If you wish to add or remove any projects, please contact us at [email protected].