dean0x7d / Pybinding
Labels
Projects that are alternatives of or similar to Pybinding
Pybinding is a Python package for numerical tight-binding calculations in solid state physics. The main features include:
-
Declarative model construction - The user just needs to describe what the model should be, but not how to build it. Pybinding will take care of the numerical details of building the Hamiltonian matrix so users can concentrate on the physics, i.e. the quantum properties of the model.
-
Fast compute - Pybinding's implementation of the kernel polynomial method allows for very fast calculation of various physical properties of tight-binding systems. Exact diagonalization is also available through the use of scipy's eigenvalue solvers. The framework is very flexible and allows the addition of user-defined computation routines.
-
Result analysis and visualization - The package contains utility functions for post-processing the raw result data. The included plotting functions are tailored for tight-binding problems to help visualize the model structure and to make sense of the results.
The code interface is written in Python with the aim to be as user-friendly and flexible as possible. Under the hood, C++11 is used to accelerate demanding tasks to deliver high performance with low memory usage.
See the documentation for more details.
Install
Pybinding can be installed on Windows, Linux or Mac, with the following prerequisites:
- Python 3.6 or newer (Python 2.x is not supported)
- The SciPy stack of scientific packages, with required versions:
- numpy >= v1.12
- scipy >= v0.19
- matplotlib >= v2.0
- If you're using Linux, you'll also need GCC >= v5.0 (or clang >= v3.5) and CMake >= v3.1.
Detailed install instructions are part of the documentation, but if you already have all the
prerequisites, it's just a simple case of using pip, Python's usual package manager:
pip install pybinding
Features
The goal of the project is to develop a tight-binding code framework which is fast, flexible and easy to use. This is just a quick overview of some of the features. See the documentation for more details and a guide to get started.
- Construction of arbitrary tight-binding lattices and geometries: 1 to 3 dimensions (including multilayer 2D systems), periodic or finite size (with fine control of edges)
- Easy polygon shape definition for 2D systems and freeform shapes for n-dimensional systems
- Geometric deformations and defects: defined via displacement and state functions
- Fields and arbitrary effects: defined via hopping and onsite energy functions
- Kernel polynomial method: a fast way to compute Green's function, spectral densities of arbitrary operators, electrical conductivity, or various other user-defined KPM methods
- Exact diagonalization: standard dense and sparse eigenvalues solvers (always available) and the FEAST solver (only available when compiled with Intel's MKL)
- Transport: scattering systems with semi-infinite leads can be constructed in pybinding and then solved using the Kwant compatibility layer
- Model and result objects have builtin plotting functions for easy visualization
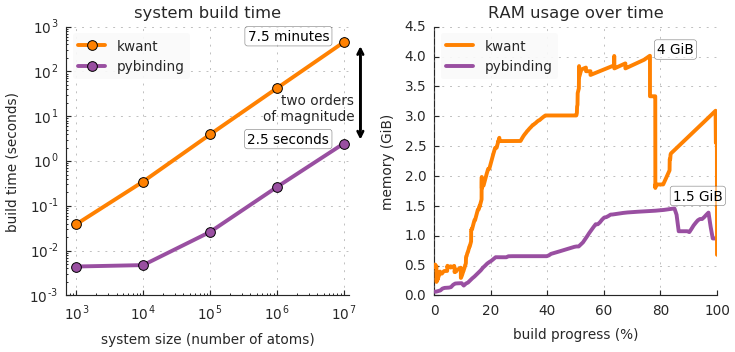
Benchmarks
One of the main features of pybinding is an easy-to-use and fast model builder. This can be a demanding task for large or complicated systems. Great care was taken to make this process fast.
The following figures compare the performance of pybinding with the Kwant package. They present the time and memory required to build a Hamiltonian matrix which describes a tight-binding system. Pybinding features good performance and a low memory footprint by using contiguous data structures and vectorized operations.
See the benchmarks section of the documentation for details on the testbed hardware and software, as well as the source code which can be used to reproduce the results.
Citing
Pybinding is free to use under the simple conditions of the BSD open source license. If you wish to use results produced with this package in a scientific publication, please just mention the package name in the text and cite the Zenodo DOI of this project:
You'll find a "Cite as" section in the bottom right of the Zenodo page. You can select a citation style from the dropdown menu or export the data in BibTeX and similar formats.
Questions?
If you have any questions, feel free to join the chat room on Gitter. You can also open an issue at the tracker.