microsoft / Recursive Cascaded Networks
Programming Languages
Labels
Projects that are alternatives of or similar to Recursive Cascaded Networks
Recursive Cascaded Networks for Unsupervised Medical Image Registration (ICCV 2019)
By Shengyu Zhao, Yue Dong, Eric I-Chao Chang, Yan Xu.
Paper link: [arXiv]
Introduction
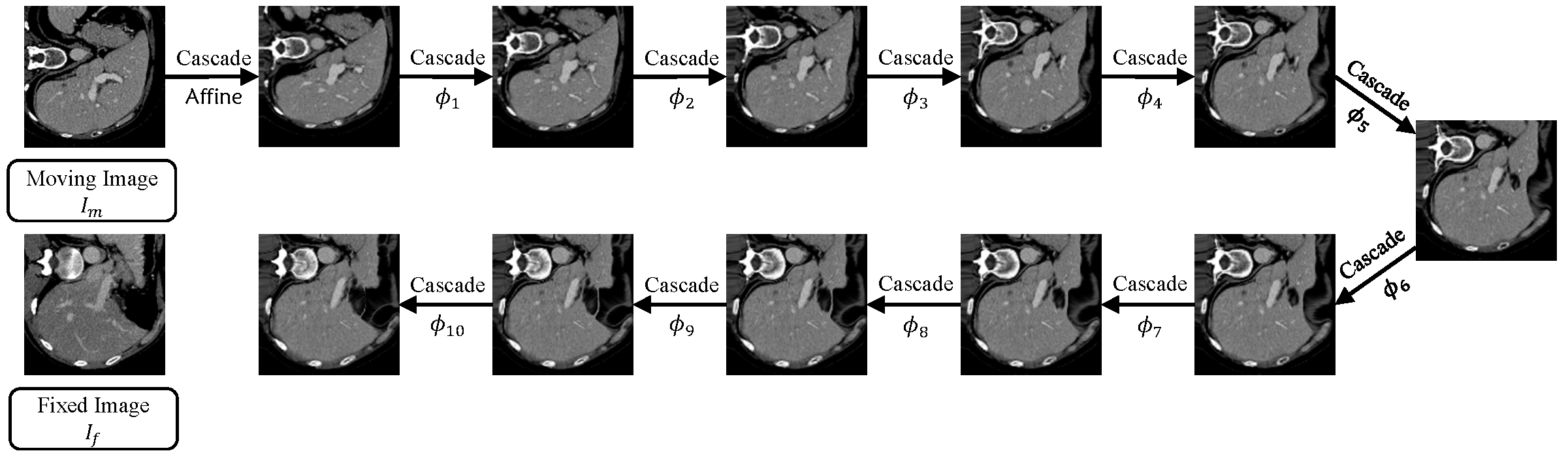
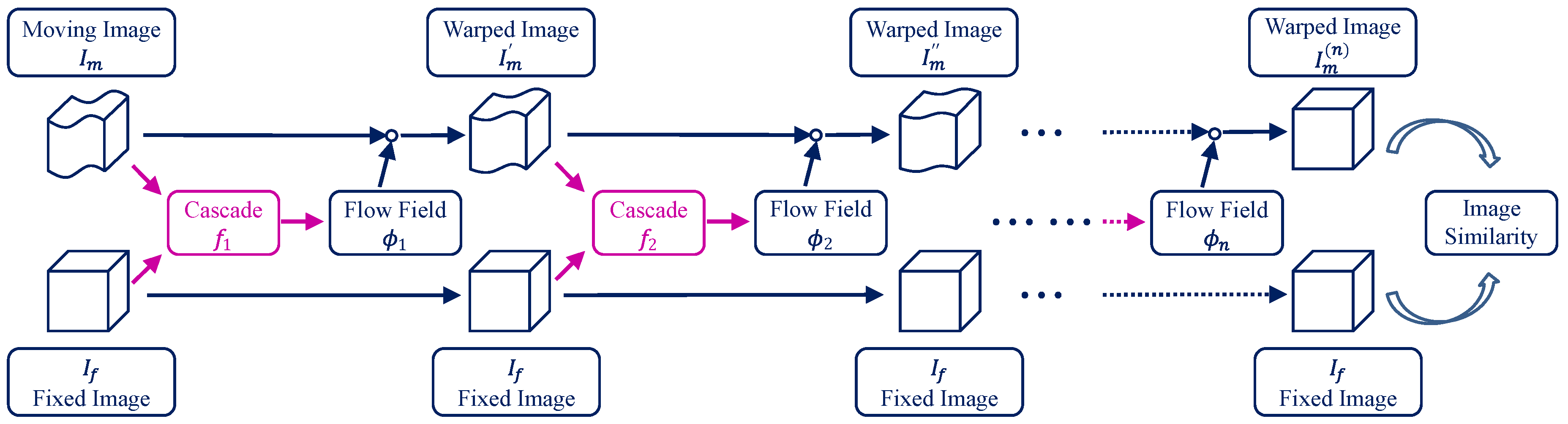
We propose recursive cascaded networks, a general architecture that enables learning deep cascades, for deformable image registration. The proposed architecture is simple in design and can be built on any base network. The moving image is warped successively by each cascade and finally aligned to the fixed image; this procedure is recursive in a way that every cascade learns to perform a progressive deformation for the current warped image. The entire system is end-to-end and jointly trained in an unsupervised manner. Shared-weight techniques are developed in addition to the recursive architecture. We achieve state-of-the-art performance on both liver CT and brain MRI datasets for 3D medical image registration. For more details, please refer to our paper.
This repository includes:
- Training and testing scripts using Python and TensorFlow;
- Pretrained models using either VTN or VoxelMorph as the base network; and
- Preprocessed training and evaluation datasets for both liver CT scans and brain MRIs.
Code has been tested with Python 3.6 and TensorFlow 1.4.
If you use the code, the models, or our data in your research, please cite:
@inproceedings{zhao2019recursive,
author = {Zhao, Shengyu and Dong, Yue and Chang, Eric I-Chao and Xu, Yan},
title = {Recursive Cascaded Networks for Unsupervised Medical Image Registration},
booktitle = {Proceedings of the IEEE International Conference on Computer Vision (ICCV)},
year = {2019}
}
@article{zhao2019unsupervised,
title = {Unsupervised 3D End-to-End Medical Image Registration with Volume Tweening Network},
author = {Zhao, Shengyu and Lau, Tingfung and Luo, Ji and Chang, Eric I and Xu, Yan},
journal = {IEEE Journal of Biomedical and Health Informatics},
year = {2019},
doi = {10.1109/JBHI.2019.2951024}
}
Datasets
Our preprocessed evaluation datasets can be downloaded here:
- SLIVER, LiTS, and LSPIG for liver CT scans; and
- LPBA for brain MRIs.
If you wish to replicate our results, please also download our preprocessed training datasets:
- MSD and BFH for liver CT scans; and
- ADNI, ABIDE, and ADHD for brain MRIs.
Please unzip the downloaded files into the "datasets" folder. Details about the datasets and the preprocessing stage can be found in the paper.
Pretrained Models
You may download the following pretrained models and unzip them into the "weights" folder.
For liver CT scans,
For brain MRIs,
Evaluation
If you wish to evaluate the pretrained 10-cascade VTN (for liver) for example, please first make sure that the evaluation datasets for liver CT scans, SLIVER, LiTS, and LSPIG, have been placed into the "datasets" folder. For evaluation on the SLIVER dataset (20 * 19 pairs in total), please run:
python eval.py -c weights/VTN-10-liver -g YOUR_GPU_DEVICES
For evaluation on the LiTS dataset (131 * 130 pairs in total, which might be quite slow), please run:
python eval.py -c weights/VTN-10-liver -g YOUR_GPU_DEVICES -v lits
For pairwise evaluation on the LSPIG dataset (34 pairs in total), please run:
python eval.py -c weights/VTN-10-liver -g YOUR_GPU_DEVICES -v lspig --paired
YOUR_GPU_DEVICES specifies the GPU ids to use (default to 0), split by commas with multi-GPU support, or -1 if CPU only. Make sure that the number of GPUs specified evenly divides the BATCH_SIZE that can be specified using --batch BATCH_SIZE (default to 4). The proposed shared-weight cascading technique can be tested using -r TIMES_OF_SHARED_WEIGHT_CASCADES (default to 1).
When the code returns, you can find the result in "evaluate/*.txt".
Similarly, to evaluate the pretrained 10-cascade VTN (for brain) on the LPBA dataset:
python eval.py -c weights/VTN-10-brain -g YOUR_GPU_DEVICES
Please refer to our paper for details about the evaluation metrics and our experimental settings.
Training
The following script is for training:
python train.py -b BASE_NETWORK -n NUMBER_OF_CASCADES -d DATASET -g YOUR_GPU_DEVICES
BASE_NETWORK specifies the base network (default to VTN, also can be VoxelMorph). NUMBER_OF_CASCADES specifies the number of cascades to train (not including the affine cascade), default to 1. DATASET specifies the data config (default to datasets/liver.json, also can be datasets/brain.json). YOUR_GPU_DEVICES specifies the GPU ids to use (default to 0), split by commas with multi-GPU support, or -1 if CPU only. Make sure that the number of GPUs specified evenly divides the BATCH_SIZE that can be specified using --batch BATCH_SIZE (default to 4). Specify -c CHECKPOINT to start with a previous checkpoint.
Demo
We provide a demo that directly takes raw CT scans as inputs (only DICOM series supported), preprocesses them into liver crops, and generates the outputs:
python demo.py -c CHECKPOINT -f FIXED_IMAGE -m MOVING_IMAGE -o OUTPUT_DIRECTORY -g YOUR_GPU_DEVICES
Note that the preprocessing stage includes cropping the liver area using a threshold-based algorithm, which takes a couple of minutes and the correctness is not guaranteed.
Built-In Warping Operation with TensorFlow
If you wish to reduce the GPU memory usage, we implemented a memory-efficient warping operation built with TensorFlow 1.4. A pre-built installer for Windows x64 can be found here, which can be installed with pip install tensorflow_gpu-1.4.1-cp36-cp36m-win_amd64.whl. Please then specify --fast_reconstruction in the training and testing scripts to enable this feature. Otherwise, the code is using an alternative version of the warping operation provided by VoxelMorph.
Acknowledgement
We would like to acknowledge Tingfung Lau and Ji Luo for the initial implementation of VTN.