jparkhill / Tensormol
Programming Languages
Projects that are alternatives of or similar to Tensormol
►
 -Title signature by Alex Graves' handwriting LSTM https://arxiv.org/abs/1308.0850
-Title signature by Alex Graves' handwriting LSTM https://arxiv.org/abs/1308.0850
Authors:
Kun Yao ([email protected]), John Herr ([email protected]), David Toth ([email protected]), Ryker McIntyre([email protected]), Nicolas Casetti, [email protected])
Model Chemistries:
- Behler-Parrinello with electrostatics
- Many Body Expansion
- Bonds in Molecules NN
- Atomwise Forces
- Inductive Charges
Simulation Types:
- Optimizations
- Molecular Dynamics (NVE,NVT Nose-Hoover)
- Monte Carlo
- Open/Periodic Boundary Conditions
- Meta-Dynamics
- Infrared spectra by propagation
- Infrared spectra by Harmonic Approximation.
- Nudged Elastic Band
- Path integral simulations via interface with I-PI MD engine.
News:
- Did we disappear? NO! but TensorMol0.2 is being developed in a private developers branch. It'll be back here soon.
- (update 3/29/2018) TensorMol0.2 will arrive before 5/1. It adds additn'l element support, geometrical constraints, conformational search and other features.
License: GPLv3
By using this software you agree to the terms in COPYING
Installation:
- Install TensorFlow(>1.1), otherwise TensorMol is self-contained.
- Works on OSX, Ubuntu, and Windows subsystem for Linux:
git clone https://github.com/jparkhill/TensorMol.git
cd TensorMol
# If you are using python2x
sudo pip install -e .
# If you are using python3x
sudo pip3 install -e .
python test.py
Demo of training a neural network force field using TensorMol:
- Copy the training script into the tensormol folder:
cp samples/training_sample.py .Run the script:python training_sample.pyThis will train a network force field for water.
Test example for TensorMol01:
- Download our pretrained neural networks (network.tar.gz). Networks for water and molecules that only contains C, H, O, N (The file is about 6 Gigabyte. This may take a while)
- Copy the zipped trained networks file (network.tar.gz) into TensorMol folder. Unzip it. The networks should be in './networks' folder.
- Copy the test script into the tensormol folder:
cp samples/test_tensormol01.py .Run the script:python test_tensormol01.pyThe test sample contains geometry optimization, molecular dynamic, harmonic IR spectrum and realtime IR spectrum.
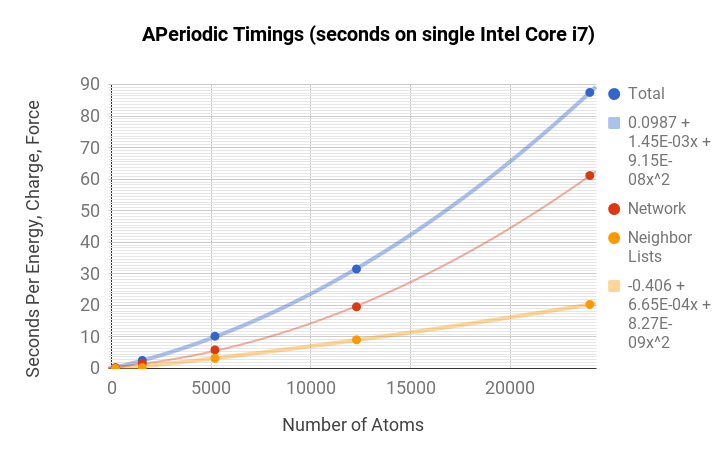
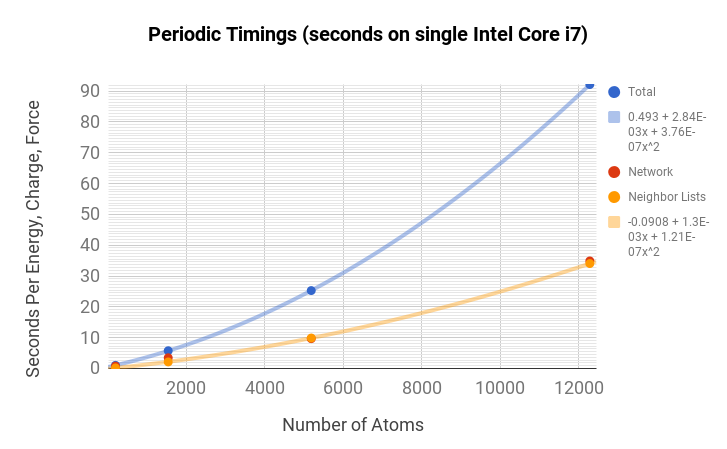
Timing Information
TensorMol is robust and fast. You can get an BP+electrostatic energy and force of this monstrous cube of 24,000 atoms in less than 100 seconds on a 2015 MacbookPro (Core i7 2.5Ghz, 16GB mem). Periodic simulations are about 3x more expensive.
Usage:
import TensorMol as tm- We are working on /doc/Tutorials, but it's sparse now. We've done a lot of re-writing, and so if you are looking for good examples, look for /samples files with recent commits.
- A collection of tests are located in samples/test_tensormol01.py but this requires first downloading the trained networks from the Arxiv paper.
python samples/test_tensormol01.py- IPI interface: start server: ~/i-pi/i-pi samples/i-pi_interface/H2O_cluster.xml > log &; run client: python test_ipi.py
Sample Results
Biological molecules
Because Neural network force fields do not rely on any specific atom typing or bond topology, the agony of setting up simulations of biological molecules is greatly reduced. This gif is a periodic optimization of PDB structure 2EVQ, in explicit polarizable TensorMol solvent.
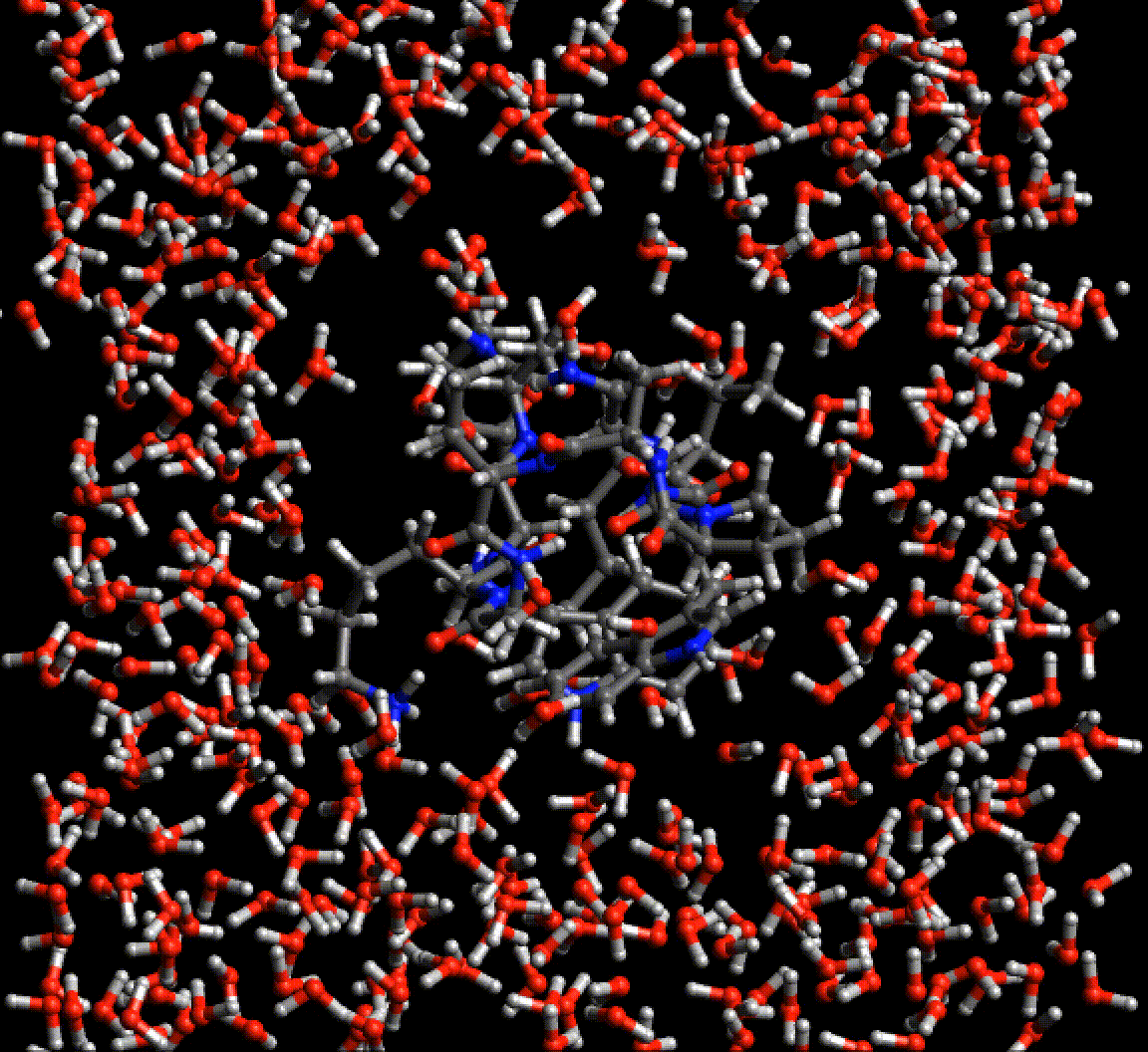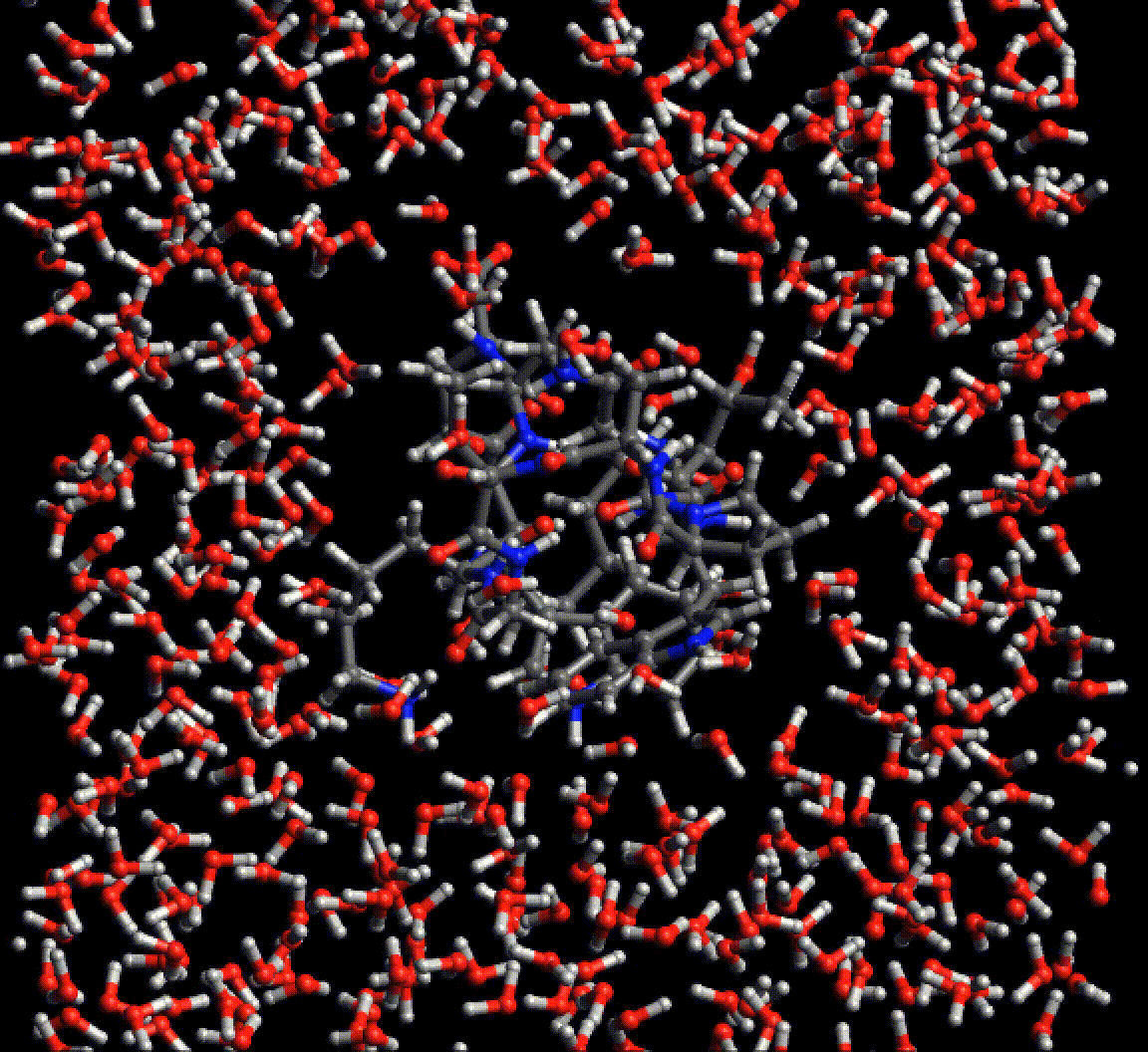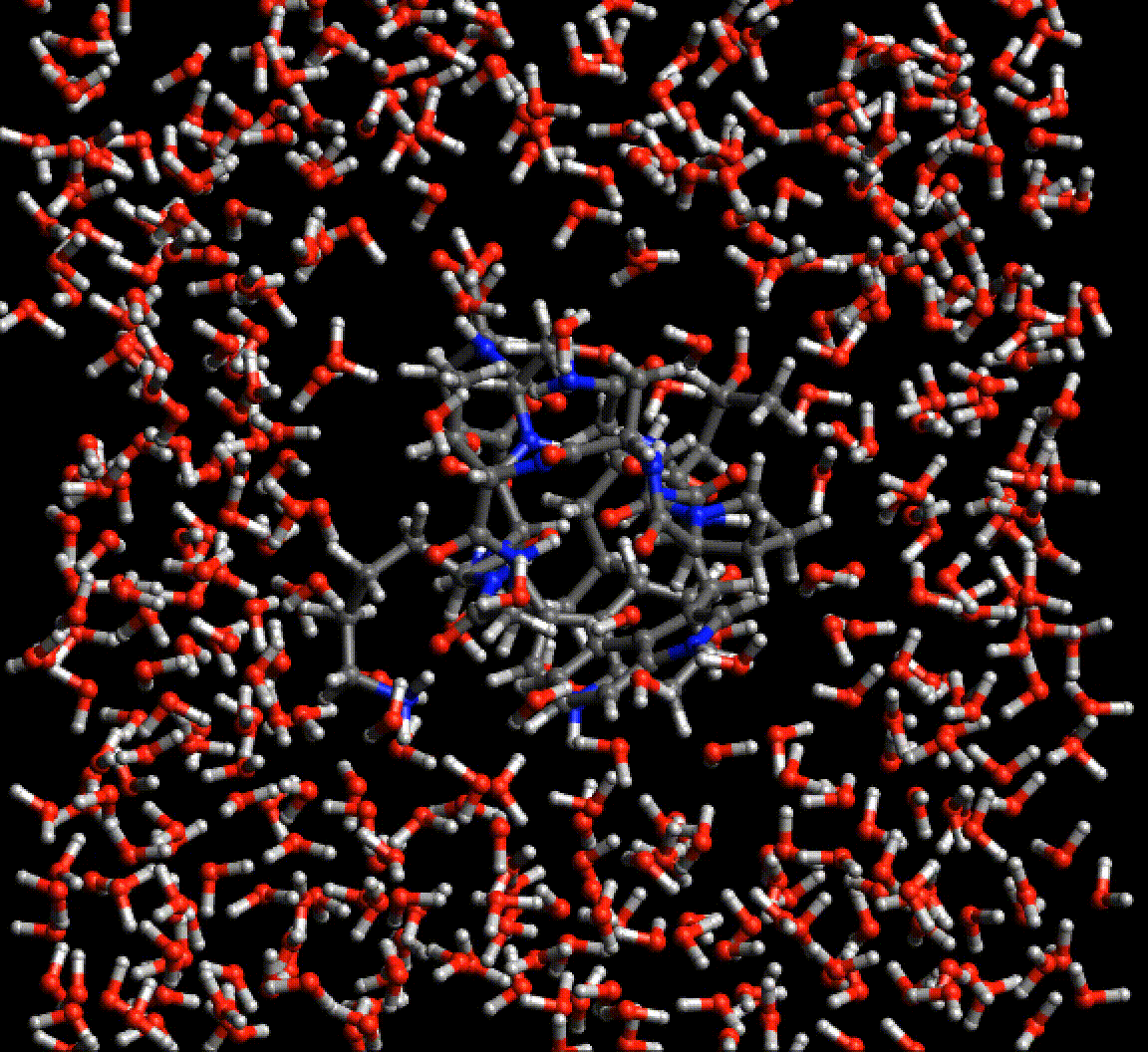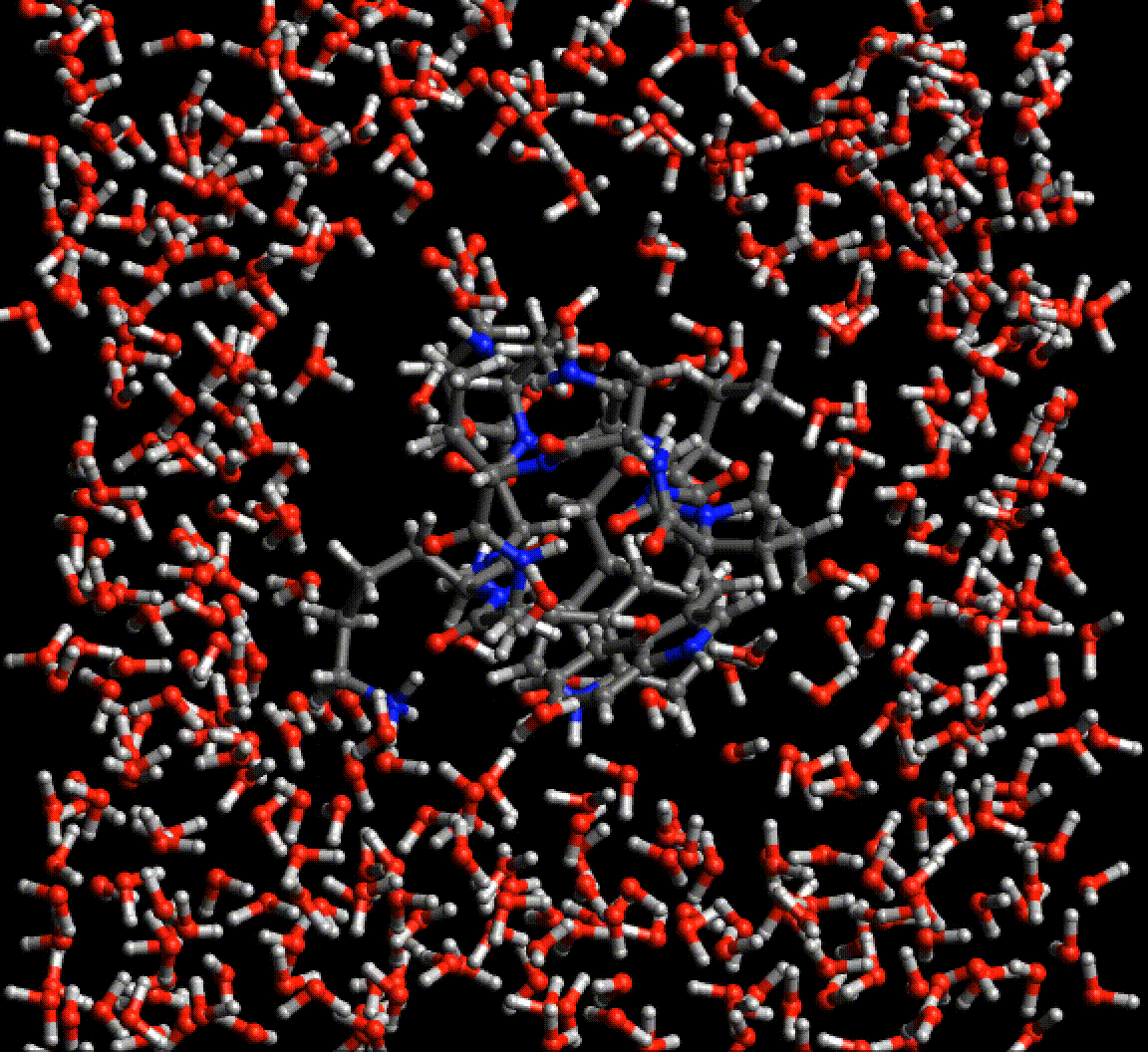
Chemical Reactions
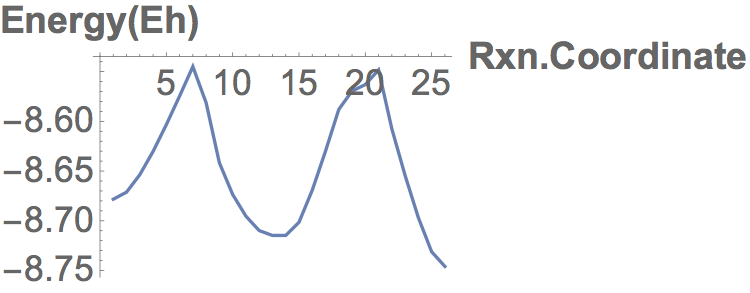
Converged nudged elastic band simulations of the cyclization cascade of endiandric acid C (c.f. K. C. Nicolaou, N. A. Petasis, R. E. Zipkin, 1982, The endiandric acid cascade. Electrocyclizations in organic synthesis. 4. Biomimetic approach to endiandric acids A-G. Total synthesis and thermal studies, J. Am. Chem. Soc. 104(20):5560–5562).
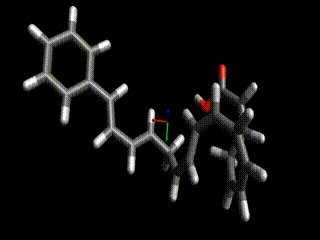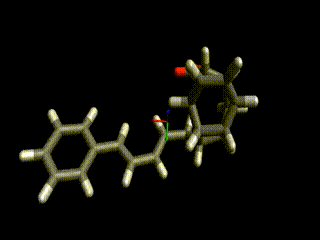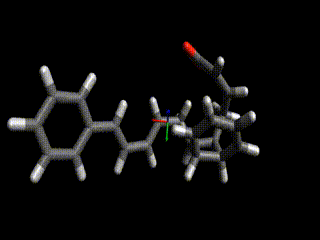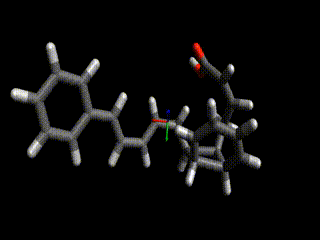
This reaction path can be found in a few minutes on an ordinary laptop. Relaxation from the linearly interpolated guess looks like this:
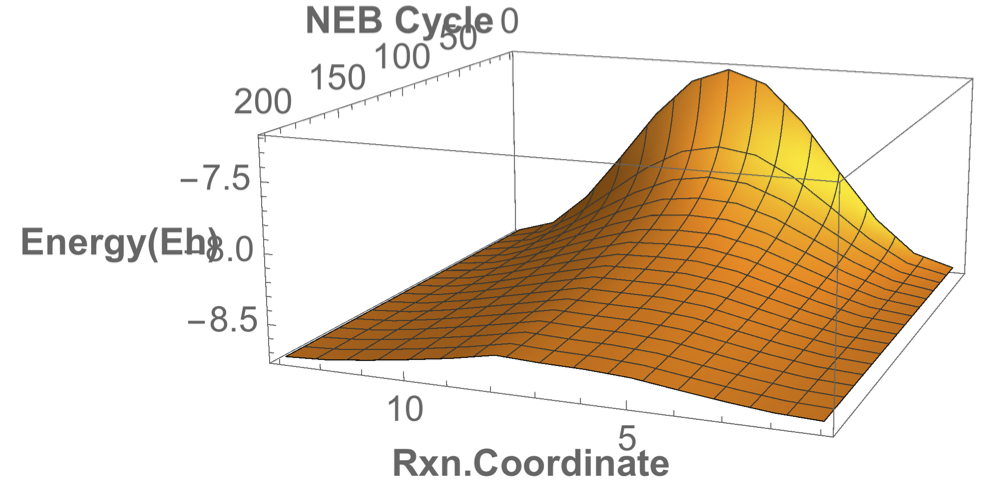
The associated energy surface is shown below.
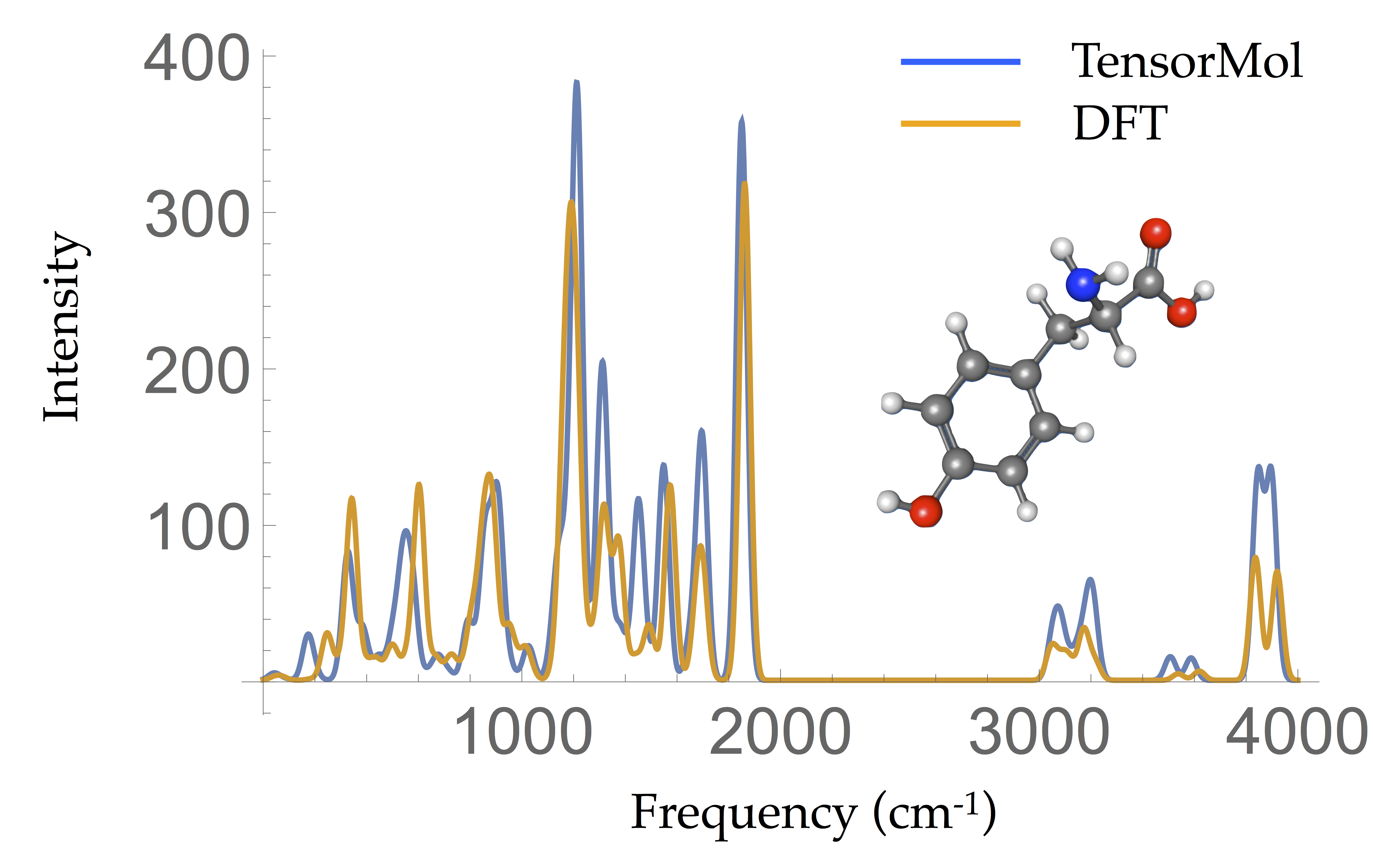
Dynamic Properties
- Tyrosine Harmonic IR spectrum
Publications and Press:
- John E. Herr, Kun Yao, David W. Toth, Ryker Mcintyre, John Parkhill.Metadynamics for Training Neural Network Model Chemistries: a Competitive Assessment. Arxiv
- Kun Yao, John E. Herr, David W. Toth, Ryker Mcintyre, John Parkhill. The TensorMol-0.1 Model Chemistry: a Neural Network Augmented with Long-Range Physics. Chemical Science
- Writeup in Chemistry World
- Kun Yao, John E. Herr, Seth N. Brown, & John Parkhill. Intrinsic Bond Energies from a Bonds-in-Molecules Neural Network. Journal of Physical Chemistry Letters (2017). DOI: 10.1021/acs.jpclett.7b01072
- Kun Yao, John Herr, & John Parkhill. The Many-body Expansion Combined with Neural Networks. Journal of Chemical Physics (2016). DOI: 10.1063/1.4973380
- Kun Yao, John Parkhill. The Kinetic Energy of Hydrocarbons as a Function of Electron Density and Convolutional Neural Networks. Journal of Chemical Theory and Computation (2016). DOI: 10.1021/acs.jctc.5b01011
Requirements:
- Minimum Pre-Requisites: Python2.7x, TensorFlow
sudo pip install tensorflow - Also now works in Python3.6.
- Useful Pre-Requisites: CUDA7.5, PySCF
- To Train Minimally: ~100GB Disk 20GB memory
- To Train Realistically: 1TB Disk, GTX1070++
- To Evaluate: Normal CPU and 10GB Mem
Acknowledgements:
- Google Inc. (for TensorFlow)
- NVidia Corp. (hardware)
- von Lilienfeld Group (for GBD9)
- Chan Group (for PySCF)
Common Issues:
- nan during training due to bad checkpoints in /networks (clean.sh)
- Also crashes when reviving networks from disk.
- if you have these issues try re-installing or:
sh clean.sh


