isayev / Ase_ani
Programming Languages
Projects that are alternatives of or similar to Ase ani
ASE-ANI
NOTICE: Python binaries built for python 3.6 and CUDA 9.2
Works only under Ubuntu variants of Linux with a NVIDIA GPU
This is a prototype interface for ANI-1x and ANI-1ccx neural network potentials for The Atomic Simulation Environment (ASE). Current ANI-1x and ANI-1ccx potentials provide predictions for the CHNO elements. The original ANI-1 and ANI-1x potentials are available in the "deprecated_original" original branch. For best performance the ANI-1x and ANI-1ccx ensembles in this branch should be used in any application.
REQUIREMENTS:
- Python 3.6 (we recommend Anaconda distribution)
- Modern NVIDIA GPU, compute capability 5.0 of newer.
- CUDA 9.2
- ASE
- MOPAC2012 or MOPAC2016 for some examples to compare results (Optional)
Installation
Clone this repository into desired folder and add environmental variables from bashrc_example.sh to your .bashrc.
To test the code run the python script: examples/ani_quicktest.py
Computed energies from the quick test on a working installation are (eV):
Initial Energy: -2078.502822821320
Final Energy: -2078.504266011399
For use cases please refer to examples folder with several iPython notebooks
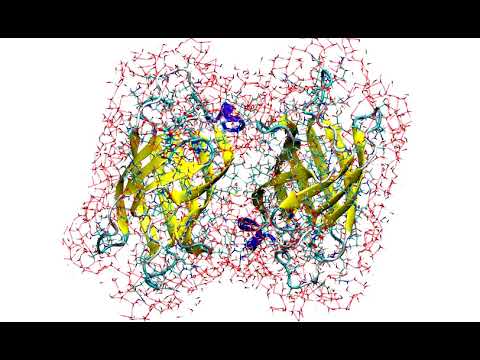
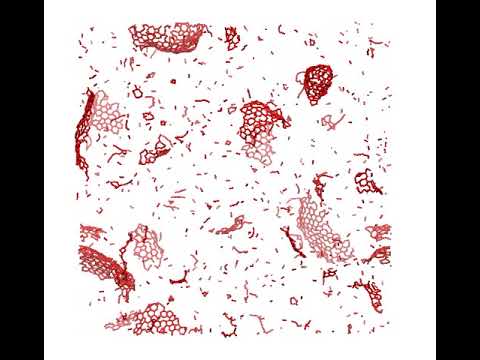
Cool stuff
Teaser of the new ANI-2x (CHNOSFCl) potential in action!
ANI-1x running 5ns MD on a box of C2 at high temperature.
ANI-1 dataset
https://github.com/isayev/ANI1_dataset
COMP6 benchmark
https://github.com/isayev/COMP6
TorchANI
We now have a PyTorch implementation. See: Documents and GitHub
Citation
If you use this code, please cite:
ANAKIN-ME ML Potential Method:
Justin S. Smith, Olexandr Isayev, Adrian E. Roitberg. ANI-1: An extensible neural network potential with DFT accuracy at force field computational cost. Chemical Science,(2017), DOI: 10.1039/C6SC05720A
Original ANI-1 data:
Justin S. Smith, Olexandr Isayev, Adrian E. Roitberg. ANI-1, A data set of 20 million calculated off-equilibrium conformations for organic molecules. Scientific Data, 4 (2017), Article number: 170193, DOI: 10.1038/sdata.2017.193 https://www.nature.com/articles/sdata2017193
Active learning-based (ANI-1x):
Justin S. Smith, Ben Nebgen, Nicholas Lubbers, Olexandr Isayev, Adrian E. Roitberg. Less is more: sampling chemical space with active learning. The Journal of Chemical Physics 148, 241733 (2018), (https://aip.scitation.org/doi/abs/10.1063/1.5023802)
Active learning and transfer learning-based (ANI-1ccx):
Justin S. Smith, Benjamin T. Nebgen, Roman Zubatyuk, Nicholas Lubbers, Christian Devereux, Kipton Barros, Sergei Tretiak, Olexandr Isayev, Adrian Roitberg. Outsmarting Quantum Chemistry Through Transfer Learning. ChemRxiv, 2018, DOI: [https://doi.org/10.26434/chemrxiv.6744440.v1]