cryptocopycats / Copycats
Programming Languages
Labels
Projects that are alternatives of or similar to Copycats
Crypto Copycats Collectibles - Buy! Sell! Hodl! Sire!
Copycats Database Tools
-
copycats - incl.
kittycommand line tool to (auto-)read kitty data records (in comma-separated values (CSV)) into an in-memory SQLite database and print reports - bitcat - bit catalog kitty browser; browse your (digital) bit(s) collections
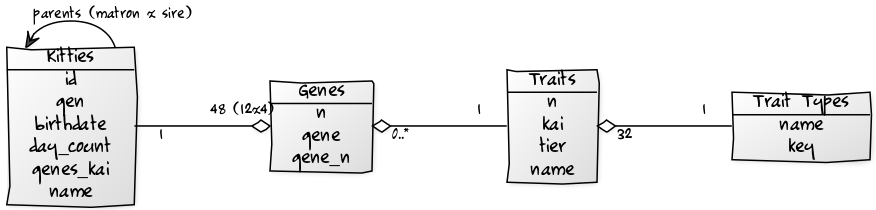
Database Tables
Table Diagram
SQL Tables (in SQLite Dialect)
CREATE TABLE kitties (
id INTEGER PRIMARY KEY AUTOINCREMENT
NOT NULL,
name VARCHAR,
genes_kai VARCHAR NOT NULL,
gen INTEGER NOT NULL,
birthdate DATETIME NOT NULL,
day_count INTEGER NOT NULL,
matron_id INTEGER,
sire_id INTEGER,
body_id INTEGER NOT NULL,
pattern_id INTEGER NOT NULL,
coloreyes_id INTEGER NOT NULL,
eyes_id INTEGER NOT NULL,
color1_id INTEGER NOT NULL,
color2_id INTEGER NOT NULL,
color3_id INTEGER NOT NULL,
wild_id INTEGER NOT NULL,
mouth_id INTEGER NOT NULL
);
CREATE TABLE genes (
id INTEGER PRIMARY KEY AUTOINCREMENT
NOT NULL,
kitty_id INTEGER NOT NULL,
n INTEGER NOT NULL,
gene VARCHAR NOT NULL,
gene_n INTEGER NOT NULL,
trait_id INTEGER NOT NULL
);
CREATE TABLE traits (
id INTEGER PRIMARY KEY AUTOINCREMENT
NOT NULL,
trait_type_id INTEGER NOT NULL,
name VARCHAR NOT NULL,
n INTEGER NOT NULL,
kai VARCHAR NOT NULL,
tier INTEGER
);
CREATE TABLE trait_types (
id INTEGER PRIMARY KEY AUTOINCREMENT
NOT NULL,
name VARCHAR NOT NULL,
[key] VARCHAR NOT NULL
);
Database Setup
Use the kitty setup command to setup an SQLite database and (auto-)read all datafiles. Example:
$ kitty setup
This will create:
- a single-file SQLite database
kitties.db - setup all tables
- add all known traits and trait types (body, pattern, eyes, ...) and
- (auto-)read all datafiles (
**/*.csv) in the.and all subdirectories
Note: Use the -i/--include option to change the default data directory (that is, .)
and use the -n/--dbname option to change the default SQLite database name (that is, kitties.db)
and use the -d/--dbpath option to change the default SQLite database path (that is, .).
Showtime! Use the sqlite3 command line tool and try some queries. Example:
$ sqlite3 kitties.db
sqlite> SELECT * FROM kitties WHERE id = 1;
1||ccac 7787 fa7f afaa 1646 7755 f9ee 4444 6766 7366 cccc eede|0|2017-11-23 06:19:59|...
sqlite> SELECT * FROM genes WHERE trait_id = 14; -- sphynx (14)
1|1|0|d|0|14
3|1|2|r2|2|14
4|1|3|r3|3|14
38|2|1|r1|1|14
146|5|1|r1|1|14
181|6|0|d|0|14
183|6|2|r2|2|14
...
Database Queries
SQL
Find all kitties with a trait
Let's use the trait savannah (fur) with the id 0:
SELECT id FROM kitties WHERE body_id = 0
Find all kitties with two traits
Let's use the trait savannah (fur) with the id 0 and the trait tiger (pattern) with the id 33:
SELECT id FROM kitties
WHERE body_id = 0 AND pattern_id = 33
Find all kitties with a trait (in any gene d/r1/r2/r3)
Note: All traits (12 x 32 = 384) are numbered with ids from 0 to 383 in the traits database table. Let's use the trait savannah (fur) with the id 0:
SELECT kitty_id FROM genes WHERE trait_id = 0
Find all kitties with a dominant (visible) trait
Note: Use gene column (d/r1/r2/r3) or the numeric gene_n
column (0/1/2/3): Let's use the trait savannah (fur) with the id 0
and a dominant (d) gene:
SELECT kitty_id FROM genes
WHERE trait_id = 0 AND gene='d'
Find all kitties with two traits (in any gene d/r1/r2/r3)
Use two query with "intersect" the result. Let's use the trait savannah (fur) with the id 0 and the trait tiger (pattern) with the id 33:
SELECT kitty_id FROM genes WHERE trait_id = 0
INTERSECT
SELECT kitty_id FROM genes WHERE trait_id = 33
Using Models w/ ActiveRecord in Ruby
Find all kitties with a trait
Let's use the trait savannah (fur) with the id 0:
Kitty.find_by( body: Trait.find_by( name: 'savannah' ))
# -or -
Kitty.find_by( body_id: 0)
Find all kitties with two traits
Let's use the trait savannah (fur) with the id 0 and the trait tiger (pattern) with the id 33:
Kitty.find_by( body: Trait.find_by( name: 'savannah' ),
pattern: Trait.find_by( name: 'tiger' ))
# -or -
Kitty.find_by( body_id: 0, pattern_id: 33 )
Find all kitties with a trait (in any gene d/r1/r2/r3)
Let's use the trait savannah (fur) with the id 0:
genes = Gene.find_by( trait: Trait.find_by( name: 'savannah' )) # query
#-or-
genes = Gene.find_by( trait_id: 0 )
genes.map { |gene| gene.kitty } # get kitties (from gene)
Find all kitties with a dominant (visible) trait
Let's use the trait savannah (fur) with the id 0 and a dominant (d) gene:
genes = Gene.find_by( trait: Trait.find_by( name: 'savannah' ),
d: 'd' ) #query
#-or-
genes = Gene.find_by( trait_id: 0, d: 'd' )
genes.map { |gene| gene.kitty } # get kitties (from gene)
Find all kitties with two traits
Use two query with "intersect" the result. Let's use the trait savannah (fur) and the trait tiger (pattern):
genes = Gene.select('kitty_id').where( trait: Trait.find_by( name: 'savannah' )).intersect(
Gene.select('kitty_id').where( trait: Trait.find_by( name: 'pattern' )))
genes.map { |gene| gene.kitty } # get kitties (from gene)
Datasets
(Crypto) Kitties on the Blockchain -
public dataset in comma-separated values (CSV) format in blocks of a thousand kitties each (e.g.
000.csv incl. 1-999,
001.csv incl. 1000-1999,
002.csv incl. 2000-2999,
and so on). The data records for kitties incl. id, gen(eration), matron+sire ids, birthdate, 48 (12x4) genes in kai (base32) notation, and more.
Add your dataset here!