biomedia-mira / Deepscm
Projects that are alternatives of or similar to Deepscm
Deep Structural Causal Models for Tractable Counterfactual Inference
This repository contains the code for the paper
N. Pawlowski+, D. C. Castro+, B. Glocker. Deep Structural Causal Models for Tractable Counterfactual Inference. Advances in Neural Information Processing Systems. 2020 [NeurIPS Proceedings] [arXiv] [NeurIPS][Poster]
(+: joint first authors)
If you use these tools or datasets in your publications, please consider citing the accompanying paper with a BibTeX entry similar to the following:
@inproceedings{pawlowski2020dscm,
author = {Pawlowski, Nick and Castro, Daniel C. and Glocker, Ben},
title = {Deep Structural Causal Models for Tractable Counterfactual Inference},
year = {2020},
booktitle={Advances in Neural Information Processing Systems},
}
Please refer to the tagged code for the code used for the NeurIPS publication.
Structure
This repository contains code and assets structured as follows:
-
deepscm/: contains the code used for running the experiments-
arch/: model architectures used in experiments -
datasets/: script for dataset generation and data loading used in experiments -
distributions/: implementations of useful distributions or transformations -
experiments/: implementation of experiments -
morphomnist/: soft link to morphomnist tools in submodules -
submodules/: git submodules
-
-
assets/-
data/:-
morphomnist/: used synthetic morphomnist dataset -
ukbb/: subset of the ukbb testset that was used for the counterfactuals
-
-
models/: checkpoints of the trained models
-
Requirements
We use Python 3.7.2 for all experiments and you will need to install the following packages:
pip install numpy pandas pyro-ppl pytorch-lightning scikit-image scikit-learn scipy seaborn tensorboard torch torchvision
or simply run pip install -r requirements.txt.
You will also need to sync the submodule: git submodule update --recursive --init.
Usage
We assume that the code is executed from the root directory of this repository.
Morpho-MNIST
You can recreate the data using the data creation script as:
python -m deepscm.datasets.morphomnist.create_synth_thickness_intensity_data --data-dir /path/to/morphomnist -o /path/to/dataset
where /path/to/morphomnist refers to the directory containing the files from the original MNIST dataset with the original morphometrics from Morpho-MNIST dataset. Alternatively we provide the generated data in data/morphomnist. You can then train the models as:
python -m deepscm.experiments.morphomnist.trainer -e SVIExperiment -m {IndependentVISEM, ConditionalDecoderVISEM, ConditionalVISEM} --data_dir /path/to/data --default_root_dir /path/to/checkpoints --decoder_type fixed_var {--gpus 0}
where IndependentVISEM is the independent model, ConditionalDecoderVISEM is the conditional model and ConditionalVISEM is the full model. The checkpoints are saved in /path/to/checkpoints or the provided checkpoints can be used for testing and plotting:
python -m deepscm.experiments.morphomnist.tester -c /path/to/checkpoint/version_?
where /path/to/checkpoint/version_? refers to the path containing the specific pytorch-lightning run. The notebooks for plotting are situated in deepscm/experiments/plotting/morphomnist.
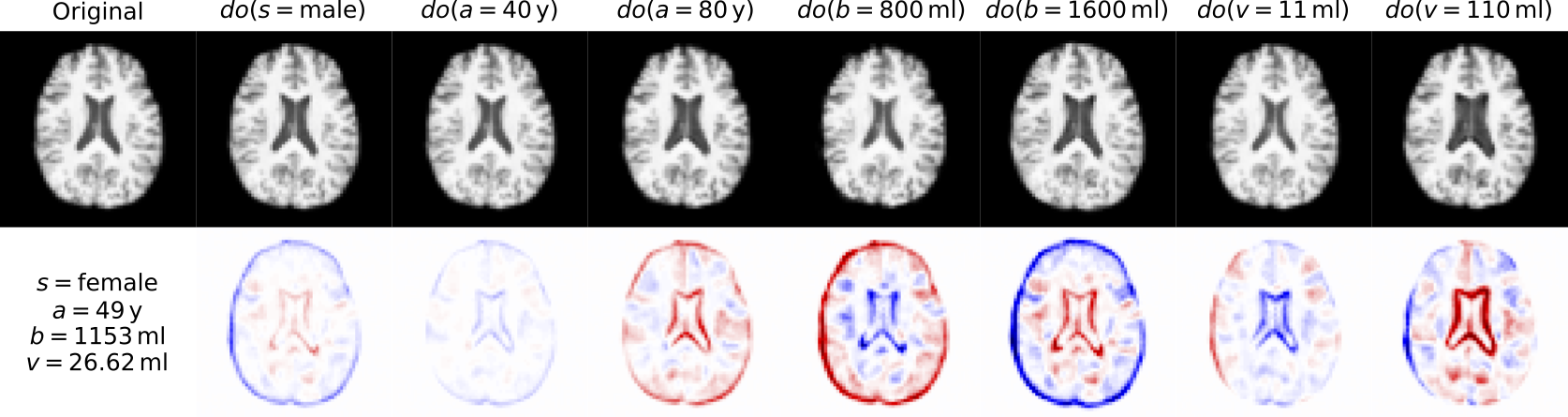
UKBB
We are unable to share the UKBB dataset. However, if you have access to the UK Biobank or a similar dataset of brain scans, you can then train the models as:
python -m deepscm.experiments.medical.trainer -e SVIExperiment -m ConditionalVISEM --default_root_dir /path/to/checkpoints --downsample 3 --decoder_type fixed_var --train_batch_size 256 {--gpus 0}
The checkpoints are saved in /path/to/checkpoints or the provided checkpoints can be used for testing and plotting:
python -m deepscm.experiments.medical.tester -c /path/to/checkpoint/version_?
The notebooks for plotting are situated in deepscm/experiments/plotting/ukbb.