titipata / Detecting Scientific Claim
Programming Languages
Projects that are alternatives of or similar to Detecting Scientific Claim
Claim Extraction for Scientific Publications
Detecting claim from scientific publication using discourse model and transfer learning. Models are trained using AllenNLP library.
Installing as a package
You can install the package using PIP, which will help you use the discourse classes inside a module
pip install git+https://github.com/titipata/detecting-scientific-claim.git
you will be able to use them as
import discourse
predictor = discourse.DiscourseCRFClassifierPredictor()
Training discourse model
Running AllenNLP to train a discourse model using PubmMedRCT dataset as follows
allennlp train experiments/pubmed_rct.json -s output --include-package discourse
We point data location to Amazon S3 directly in pubmed_rct.json
so you do not need to download the data locally. Change cuda_device to -1 in pubmed_rct.json
if you want to run on CPU. There are more experiments available in experiments folder.
Note that you have to remove output folder first before running.
Predicting discourse
We trained the Bidirectional LSTM model on structured abstracts from Pubmed to predict
discourse probability (RESULTS, METHODS, CONCLUSIONS, BACKGROUND, OBJECTIVE)
of a given sentence. You can download trained model from Amazon S3
wget https://s3-us-west-2.amazonaws.com/pubmed-rct/model.tar.gz # or model_crf.tar.gz for pretrained model with CRF layer
and run web service for discourse prediction task as follow
bash web_service.sh
To test the train model with provided examples fixtures.json,
simply run the following to predict labels.
allennlp predict model.tar.gz \
pubmed-rct/PubMed_200k_RCT/fixtures.json \
--include-package discourse \
--predictor discourse_predictor
or run the following for
allennlp predict model_crf.tar.gz \
pubmed-rct/PubMed_200k_RCT/fixtures_crf.json \
--include-package discourse \
--predictor discourse_crf_predictor
To evaluate discourse model, you can run the following command
allennlp evaluate model.tar.gz \
https://s3-us-west-2.amazonaws.com/pubmed-rct/test.json \
--include-package discourse
Predicting claim (web service)
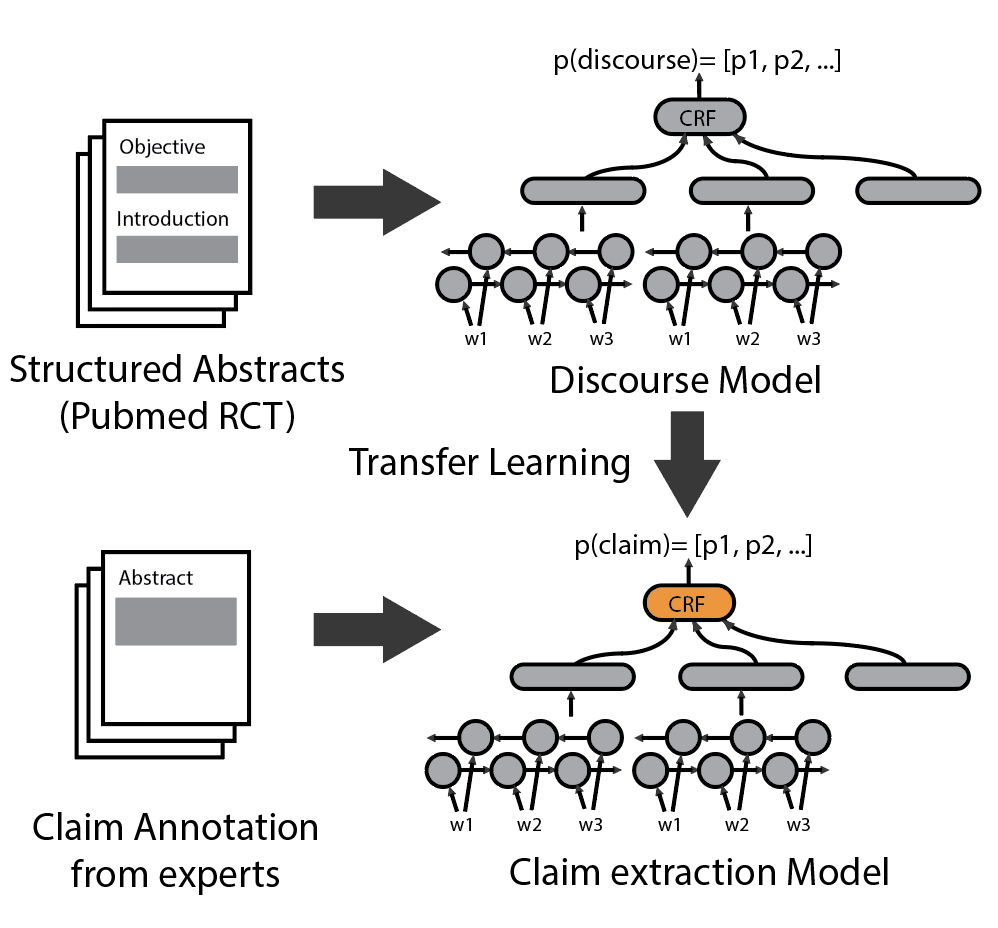
We use transfer learning with fine tuning to train claim extraction model from pre-trained discourse model. The schematic of the training can be seen below.
You can run the demo web application to detect claims as follows
export FLASK_APP=main.py
flask run --host=0.0.0.0 # this will serve at port 5000
The interface will look something like this
And output will look something like the following (highlight means claim, tag behind the sentence is discourse prediction)
Expertly annotated dataset We release the dataset of annotated 1,500 abstracts containing 11,702 sentences (2,276 annotated as claim sentences) sampled from 110 biomedical journals. The final dataset are the majority vote from three experts. The annotations are hosted on Amazon S3 and can be found from these given URLs.
Requirements
- Python 3.6
- AllenNLP >= 0.6.1
- spacy
- fastText
- Pubmed RCT - dataset
Citing the repository
You can cite our paper available on arXiv as
Achakulvisut, Titipat, Chandra Bhagavatula, Daniel Acuna, and Konrad Kording. "Claim Extraction in Biomedical Publications using Deep Discourse Model and Transfer Learning." arXiv preprint arXiv:1907.00962 (2019).
or using BibTeX
@article{achakulvisut2019claim,
title={Claim Extraction in Biomedical Publications using Deep Discourse Model and Transfer Learning},
author={Achakulvisut, Titipat and Bhagavatula, Chandra and Acuna, Daniel and Kording, Konrad},
journal={arXiv preprint arXiv:1907.00962},
year={2019}
}
Acknowledgement
This project is done at the Allen Institute for Artificial Intelligence and Konrad Kording lab, University of Pennsylvania