mlin / Genomicsqlite
Projects that are alternatives of or similar to Genomicsqlite
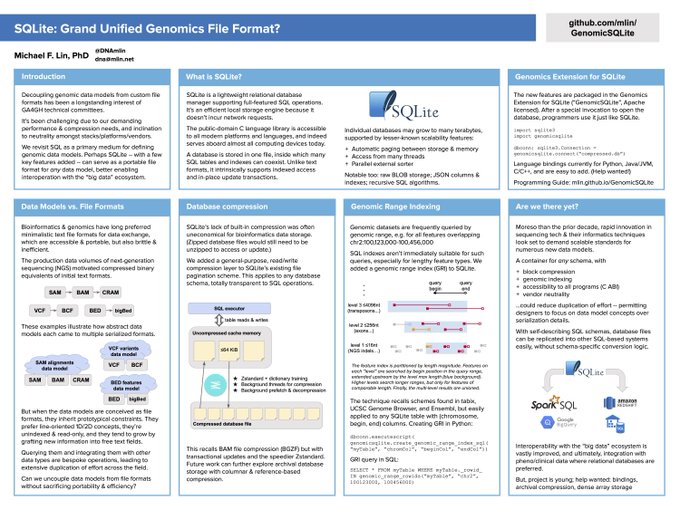
Genomics Extension for SQLite
("GenomicSQLite")
This SQLite3 loadable extension adds features to the ubiquitous embedded RDBMS supporting applications in genome bioinformatics:
- genomic range indexing for overlap queries & joins
- in-SQL utility functions, e.g. reverse-complement DNA, parse "chr1:2,345-6,789"
- automatic streaming storage compression (also available standalone)
- reading directly from HTTP(S) URLs (also available standalone)
- pre-tuned settings for "big data"
This October 2020 poster discusses the context and long-run ambitions:
Our Colab notebook demonstrates key features with Python, one of several language bindings.
USE AT YOUR OWN RISK: The extension makes fundamental changes to the database storage layer. While designed to preserve ACID transaction safety, it's young and unlikely to have zero bugs. This project is not associated with the SQLite developers.
Installation & Programming Guide
Start Here 👉 full documentation site
We supply the extension prepackaged for Linux x86-64 and macOS Catalina. An up-to-date version of SQLite itself is also required, as specified in the docs.
Programming language support:
- C/C++
- Python ≥3.6
- Java & JVM languages
- Rust
More to come. (Help wanted; see Language Bindings Guide)
Building from source
Most will prefer to install a pre-built shared library (see above). To build from source, see our Actions yml (Ubuntu 20.04) or Dockerfile (CentOS 7) used to build the more-portable releases. Briefly, you'll need:
- C++11 build system
- CMake ≥ 3.14
- Dev packages: SQLite ≥ 3.31.0, Zstandard ≥ 1.3.4, libcurl
And incantations:
cmake -DCMAKE_BUILD_TYPE=Release -B build .
cmake --build build -j 4 --target genomicsqlite
...generating build/libgenomicsqlite.so. To run the test suite, you'll furthermore need:
- htslib ≥ 1.9, samtools, and tabix
- pigz
- Python ≥ 3.6 and packages: pytest pytest-xdist pre-commit black pylint flake8
- clang-format & cppcheck
to:
pre-commit run --all-files # formatters+linters
cmake -DCMAKE_BUILD_TYPE=Debug -B build .
cmake --build build -j 4
env -C build ctest -V