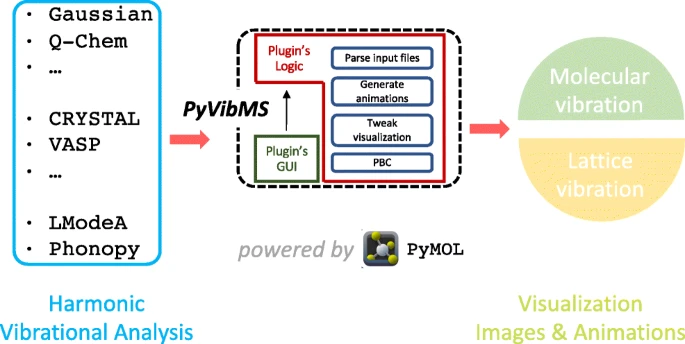
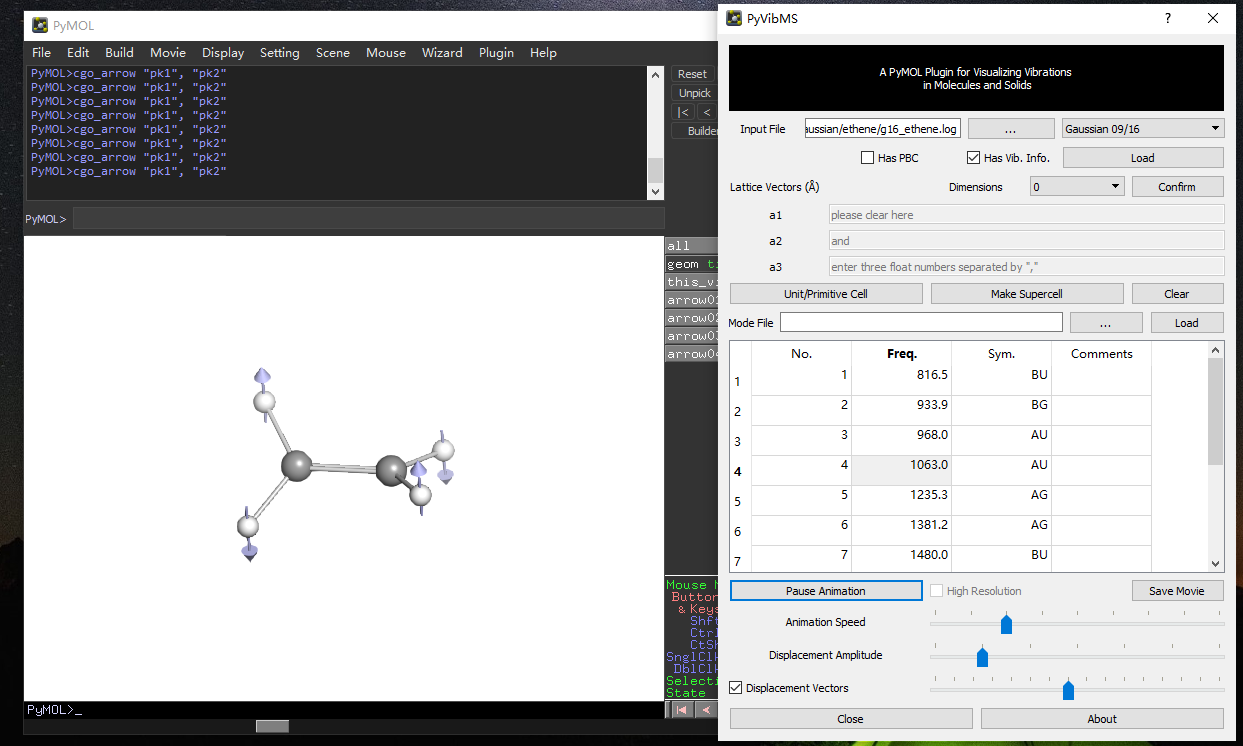
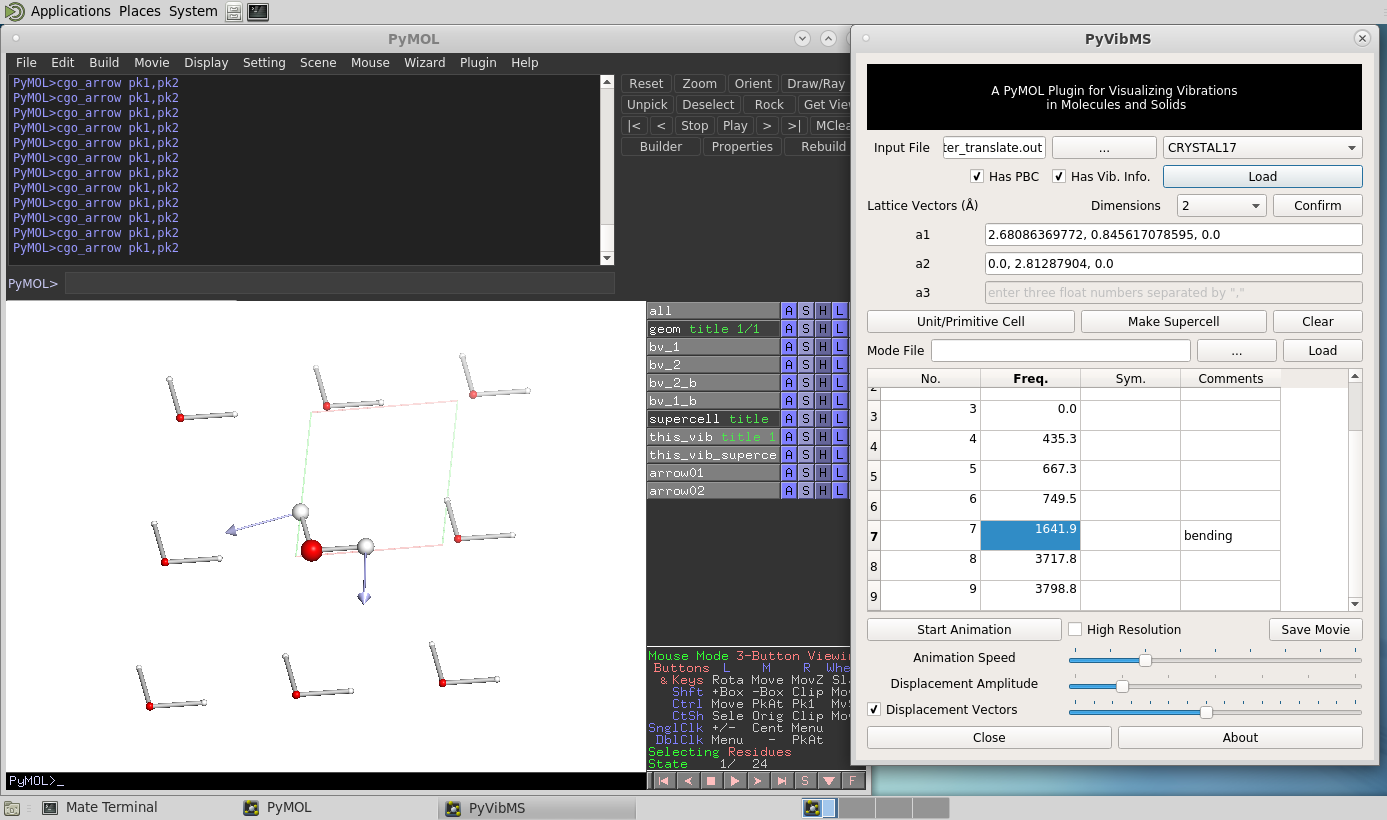
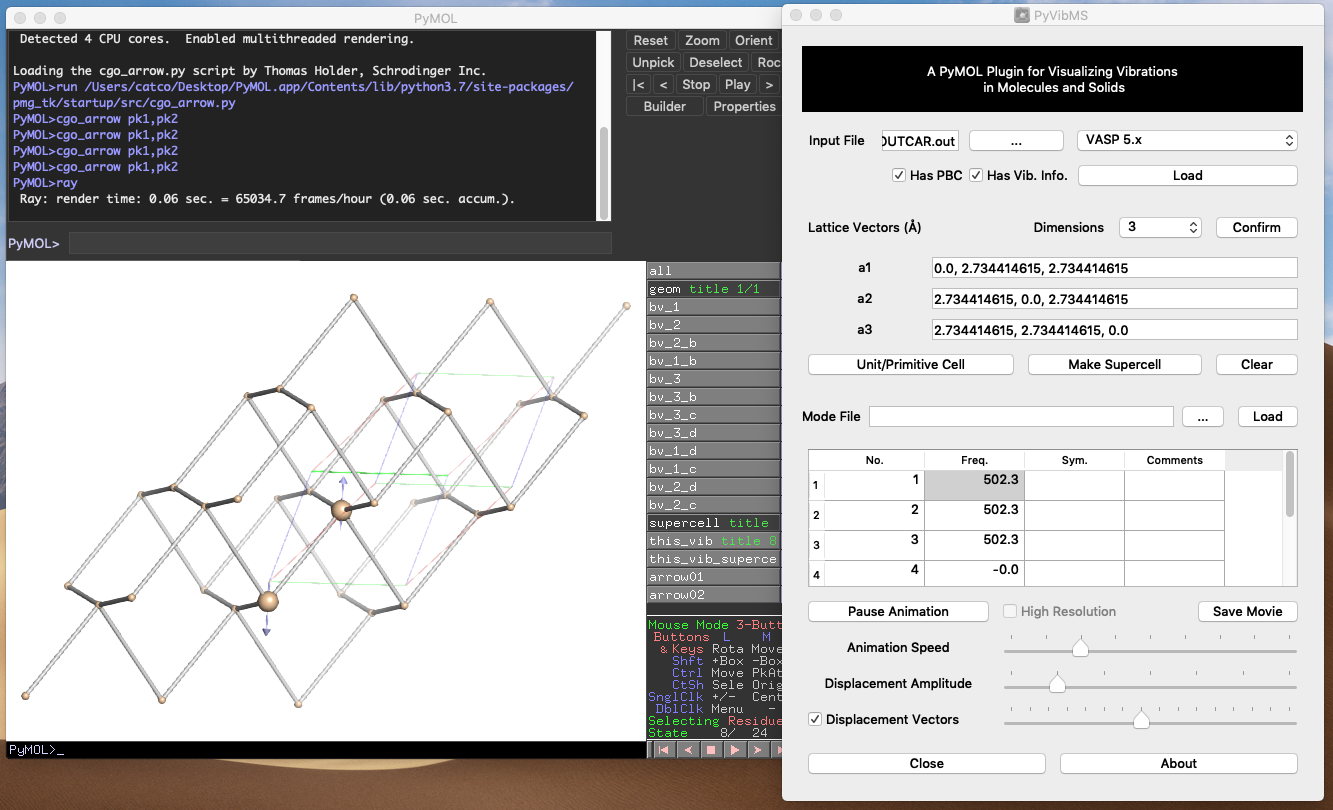
PyVibMS
A PyMOL plugin for visualizing vibrations in molecules and solids
Updates
Oct 22, 2021 - Bug fix
May 8, 2021 - Add example for BDF
May 5, 2021 - Add examples for UniMoVib which can generate XYZ and mode files for PyVibMS
May 3, 2021 - Add support for ORCA 4 and Q-Chem 4/5
May 2, 2021 - Add support for Prof. Grimme's xtb program
User Guide
介绍PyVibMS使用的中文文章 请前往 http://bbs.keinsci.com/thread-22835-1-1.html
Our paper on PyVibMS is now published.
PyVibMS: a PyMOL plugin for visualizing vibrations in molecules and solids, Journal of Molecular Modeling, 2020, 26, 290. (full text also available at https://sites.smu.edu/dedman/catco/publications/pdf/434.pdf)
This paper serves as a detailed manual of PyVibMS and a few typographical errors need to be corrected:
- Page 2
- Page 3
- Page 5
Installation
-
Please install PyMOL 2.x and download the zip file of this repository
-
Open PyMOL 2.x
-
Click "Plugin" -> "plugin manager" -> "Install New Plugin" -> "Choose", then choose the "src/__init__.py" file
-
The "PyVibMS" will be installed to PyMOL and show up in the "Plugin" drop-in menu
PyVibMS runs on various operating systems.
Windows
Linux
Mac OS
Supported quantum chemistry programs
Natively supported
- Gaussian 09/16
- ORCA 4
- xtb
- Q-Chem
- CRYSTAL17
- VASP
Generically supported
The following packages are supported with the help of UniMoVib.
- Adf
- BDF
- CFour
- Columbus
- CP2k
- Dalton
- deMon2k
- Dmol3
- Firefly
- Gamess
- Gamess-UK
- Molcas and OpenMolcas
- Molpro
- Mopac
- NWChem
- Pqs
- Psi4
- Turbomole
- Quantum ESPRESSO
- DFTB+
- and more
Other analysis tools
Tips
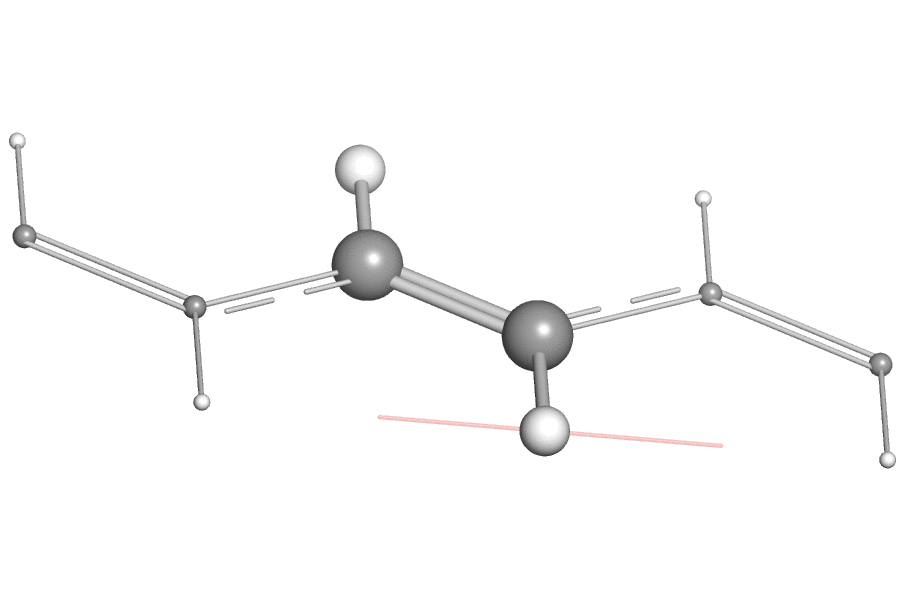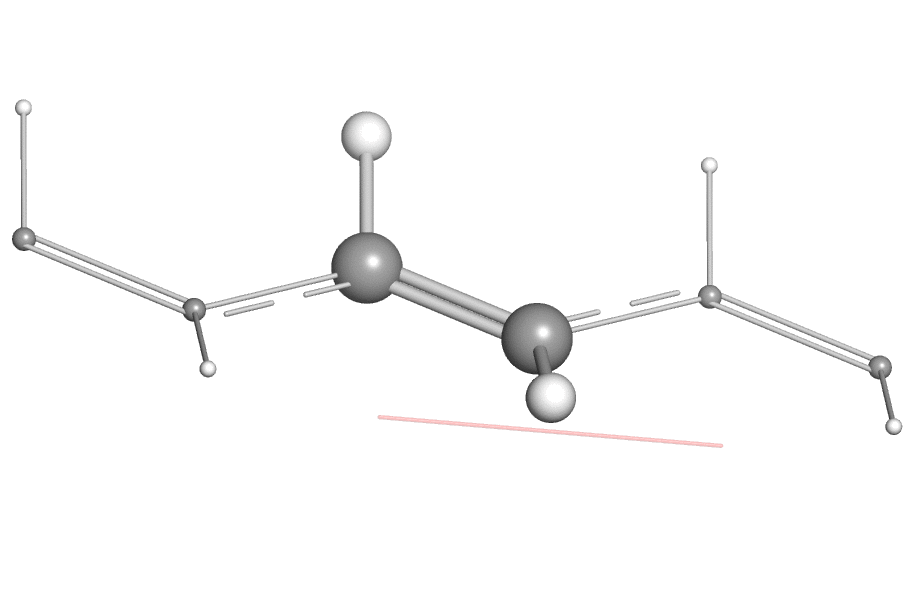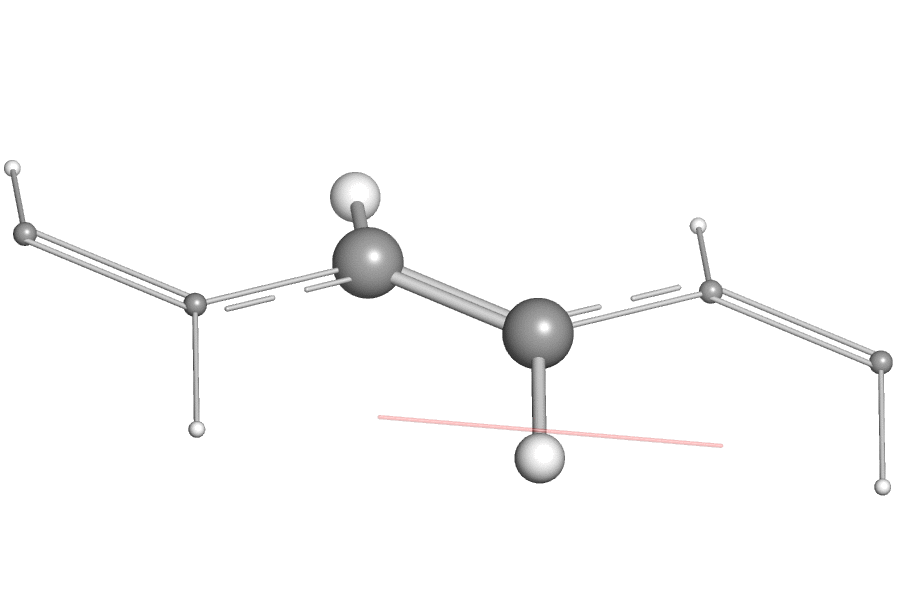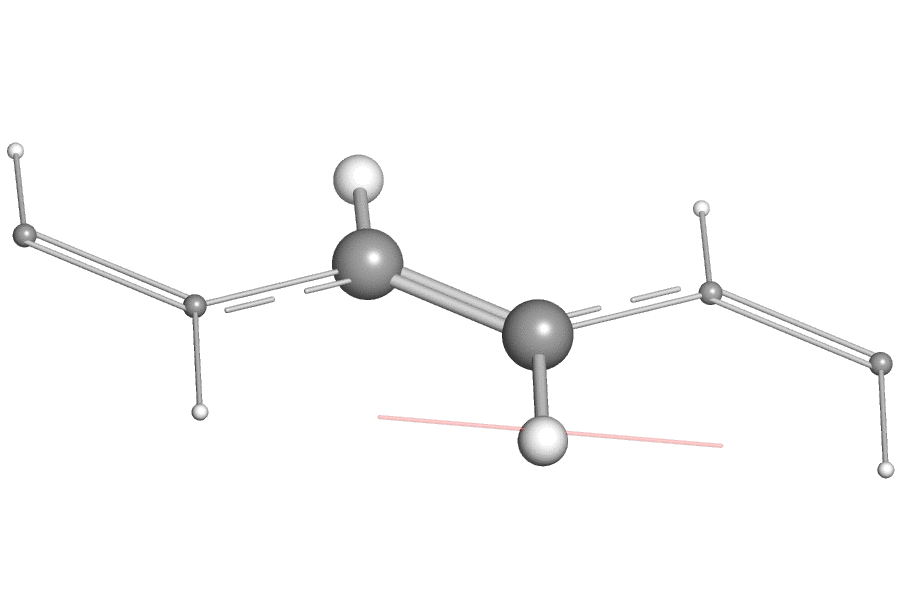
- When using the open-source version of PyMOL, one may find it difficult to export the vibration animation directly as a GIF or QuickTime movie due to the missing encoder, a work-around is to export the animation as "PNG images" (by clicking "File"->"Export Movie As"->"PNG Images..."). Then one can use a third-party tool like ezgif to combine these PNG images as animated GIF image.
Here is an example