liaohaofu / Adn
Programming Languages
Projects that are alternatives of or similar to Adn
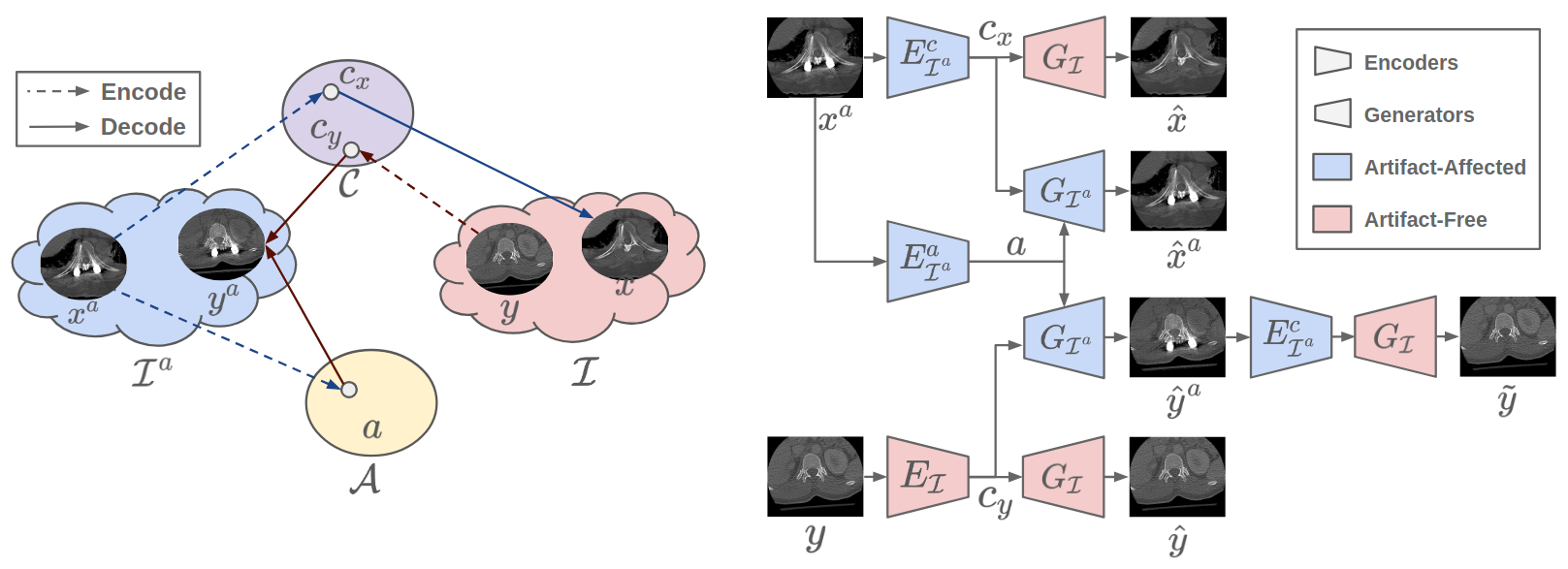
ADN: Artifact Disentanglement Network for Unsupervised Metal Artifact Reduction [Paper]
By Haofu Liao ([email protected]), Spring, 2019
Citation
If you use this code for your research, please cite our paper.
@inproceedings{adn2019_miccai,
title={Artifact Disentanglement Network for Unsupervised Metal Artifact Reduction},
author={Haofu Liao, Wei-An Lin, Jianbo Yuan, S. Kevin Zhou, Jiebo Luo},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI)},
year={2019}
}
@article{adn2019_tmi,
author={H. {Liao} and W. {Lin} and S. K. {Zhou} and J. {Luo}},
journal={IEEE Transactions on Medical Imaging},
title={ADN: Artifact Disentanglement Network for Unsupervised Metal Artifact Reduction},
year={2019},
doi={10.1109/TMI.2019.2933425}
}
Prerequisites
This repository is tested under the following system settings:
- Ubuntu 16.04
- Python 3.7 (Anaconda/Miniconda reconmmended)
- Pytorch 1.0.0 or above
- CUDA 9.0 or above
- Matlab R2018b (with Image Processing Toolbox, Parallel Computing Toolbox and Statistics and Machine Learning Toolbox)
Install
Local
For most of the users, you may consider install ADN locally on your machine with the following steps.
- Clone this repository from Github
git clone https://github.com/liaohaofu/adn.git
- Install Pytorch and Anaconda/Miniconda
- Anaconda/Miniconda installation is optional. If not installed, you may have to install some dependent python packages manually.
- Install Python dependencies.
pip install -r requirements.txt
Docker
For Docker users, we provide a pre-built docker image as well as a Dockerfile.
- Install docker-ce and nvidia-docker.
- Pull the ADN docker image from Docker Hub. This will install ADN as well as its dependencies automatically.
docker pull liaohaofu/adn
-
[Optional] If you want a customized version of ADN docker image, you may modify the docker file at
docker/Dockerfileand then build a docker image.
cd docker/
docker build -t liaohaofu/adn .
- Run the ADN docker image.
docker run -it --runtime=nvidia liaohaofu/adn
Datasets
Two publicly available datasets (DeepLesion and Spineweb) are supported. As a courtesy, we also support training/testing with natural images.
DeepLesion
- Download the DeepLesion dataset. We use the first 9 .zip files (Images_png_01.zip to Images_png_09.zip) in our experiments. You may use the
batch_download_zips.pyprovided by DeepLesion to batch download the .zip files at once. - Extract the downloaded .zip files. All the extracted images will be under the folder
path_to_DeepLesion/Images_png/. Herepath_to_DeepLesionis the folder path where you extract the .zip files. - Create a softlink to DeepLesion. Replace
path_to_DeepLesion/Images_pngto the actual path in your system before running the following command.
ln -s path_to_DeepLesion/Images_png data/deep_lesion/raw
- Prepare DeepLesion dataset for ADN (MATLAB required). The configuration file for preparing DeepLesion dataset can be found at
config/dataset.yaml.
>> prepare_deep_lesion
Spineweb
- Download the Spineweb dataset.
- Extract the spine-*.zip files (i.e.,
spine-1.zip,spine-2.zip, etc.). All the extracted images will be under the folderpath_to_Spineweb/spine-*/. Herepath_to_Spinewebis the folder path where you extract the spine-*.zip files. - Create a softlink to Spineweb. Replace
path_to_Spineweb/to the actual path to in your system before running the following command.
mkdir data/spineweb
ln -s path_to_Spineweb/ data/spineweb/raw
- Prepare Spineweb dataset for ADN. The configuration file for preparing Spineweb dataset can be found at
config/dataset.yaml.
python prepare_spineweb.py
Natural image
- Our code assumes you have prepared your natural image dataset as following.
your_dataset
├── test
│ ├── artifact # a folder containing all the testing images with artifact
│ └── no_artifact # a folder containing all the testing images without artifact
└── train
├── artifact # a folder containing all the training images with artifact
└── no_artifact # a folder containing all the training images without artifact
- Create a softlink to your natural image dataset
ln -s path_to_your_dataset data/nature_image
- Note that our model is not tuned for natural images (e.g., choices of loss functions, hyperparameters, etc.) and its effectiveness may vary depending on the problems and datasets.
Demo
- We provide a demo code to demonstrate the effectiveness of ADN. The input sample images are located at
samples/and the outputs of the demo can be found atresults/. To run the demo,
python demo.py deep_lesion
python demo.py spineweb
-
[Optional] By default, the demo code will download pretrained models from google drive automatically. If the downloading fails, you may download them from google drive manually.
- Download pretrained models from Google Drive (DeepLesion | Spineweb) or Bitbucket.
- Move the downloaded models to
runs/.
mv path_to_DeepLesion_model runs/deep_lesion/deep_lesion_49.pt mv path_to_Spineweb_model runs/spineweb/spineweb_39.pt
Train and Test
-
Configure the training and testing. We use a two-stage configuration for ADN, one for the default settings and the other for the run settings.
- The default settings of ADN can be found at
config/adn.yamlwhich is not subject to be changed. When users do not provide the values for a specific setting, the default setting in this file will be used. - The run settings can be found at
runs/adn.yaml. This is where the users provide specific settings for ADN's training and testing. Any provided settings in this file will override the default settings during the experiments. By default, the settings for training and testing ADN with DeepLesion, Spineweb and natural image datasets are provided inruns/adn.yaml.
- The default settings of ADN can be found at
-
Train ADN with DeepLesion, Spineweb datasets or a natural image dataset. The training results (model checkpoints, configs, losses, training visualizations, etc.) can be found under
runs/run_name/whererun_namecan be eitherdeep_lesion,spinewebornature_image
python train.py deep_lesion
python train.py spineweb
python train.py nature_image
- Test ADN with DeepLesion, Spineweb datasets or a natural image dataset. The testing results (evaluation metrics and testing visualizations, etc.) can be found under
runs/run_name/whererun_namecan be eitherdeep_lesion,spinewebornature_image.
python test.py deep_lesion
python test.py spineweb
python test.py nature_image
Acknowledgement
The authors would like to thank Dr. Yanbo Zhang ([email protected]) and Dr. Hengyong Yu ([email protected]) for providing the artifact synthesis code used in this repository.