dalmia / Brain Tumor Segmentation Keras
Programming Languages
Projects that are alternatives of or similar to Brain Tumor Segmentation Keras
Brain-Tumor-Segmentation-Keras
Keras implementation of the multi-channel cascaded architecture introduced in the paper "Brain Tumor Segmentation with Deep Neural Networks" by Mohammad Havaei, Axel Davy, David Warde-Farley, Antoine Biard, Aaron Courville, Yoshua Bengio, Chris Pal, Pierre-Marc Jodoin, Hugo Larochelle.
Architecture
-
Two pathways (channels) instead of the usual single feedforward. One is the local channel, looking at the local context of the pixel and the other is the global channel, looking at the global context of the image.
-
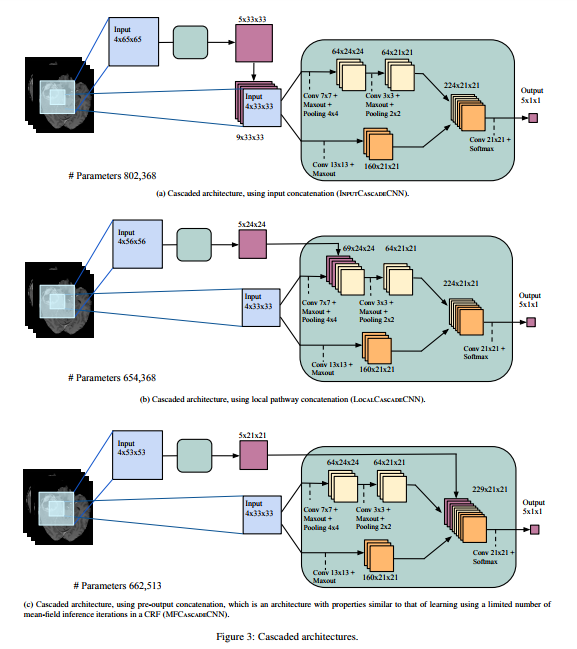
In the usual feedforward architecture, the values of the nearby pixels affect a pixel's prediction. But it makes sense that the prediction of nearby pixels should also influence a pixel's prediction. So, the paper proposes a cascaded architecture, where there are multiple choices (input, local and final) for the cascade to be applied (see figure above), leading to three possible architectures.
-
Fully connected layers replaced by 1x1 Convolutions to speed up computation.
-
2-phase training: The dataset is highly unbalanced. So, the authors propose 2-phase training, where in the 1st phase, the entire model is trained using a balanced dataset and in the 2nd phase, only the classification layer is fine-tuned according to the actual data distribution.
Building the model
# Required for Layers which have different functions for
# training / testing (e.g. Dropout, BatchNormalization)
Kc.set_learning_phase(True)
multiCascadeCNN = MultichannelCascadeCNN(mode='final')
model = multiCascadeCNN.build_model()
print(model.summary())
Dependencies
- TensorFlow
- Keras
Install them using pip.
Contributing
It would be great if someone is willing to implement the actual paper results. Feel free to create a Pull Request. If you are a beginner, you can refer this for getting started.
Support
If you found this useful, please consider starring(★) the repo so that it can reach a broader audience.