LINK TO deepflash2 and deepflash2 docs
+ **Update**
+ An improved version, deepflash2, is now available.
+ This version is no longer maintained.DeepFLaSH
Official repository of DeepFLasH - a deep learning pipeline for segmentation of fluorescent labels in microscopy images. Check out our paper on biorXiv.
Jypter Notebooks on Google Colab
- Select 'Runtime' -> 'Change runtime time' -> 'Python 3' (and 'GPU') before running the notebook.
- Ignore the warning: 'This notebook was not authored by Google.' Check 'Reset all runtimes before running' and click 'Run anyway'
- Use Firefox or Google Chrome if you want to upload your images.
Train or fine-tune your own model and segment your images
You can find a comprehensive user guide here.
Model Library
This list contains download links to the weights of the selected models as well as an example of their corresponding training images and masks.
You can select and apply these models within our Jupyter Notebook.
-
cFOS_Wue: Trained on 36 image-mask pairs of cFOS labels in the dorsal hippocampus (including 12 images of each sub-region: dentate gyrus, CA3 and CA1). Masks for training were prepared by five independent experts. Images were acquired using laser-scanning confocal microscopy with a resolution of 1.6 pixel per µm.
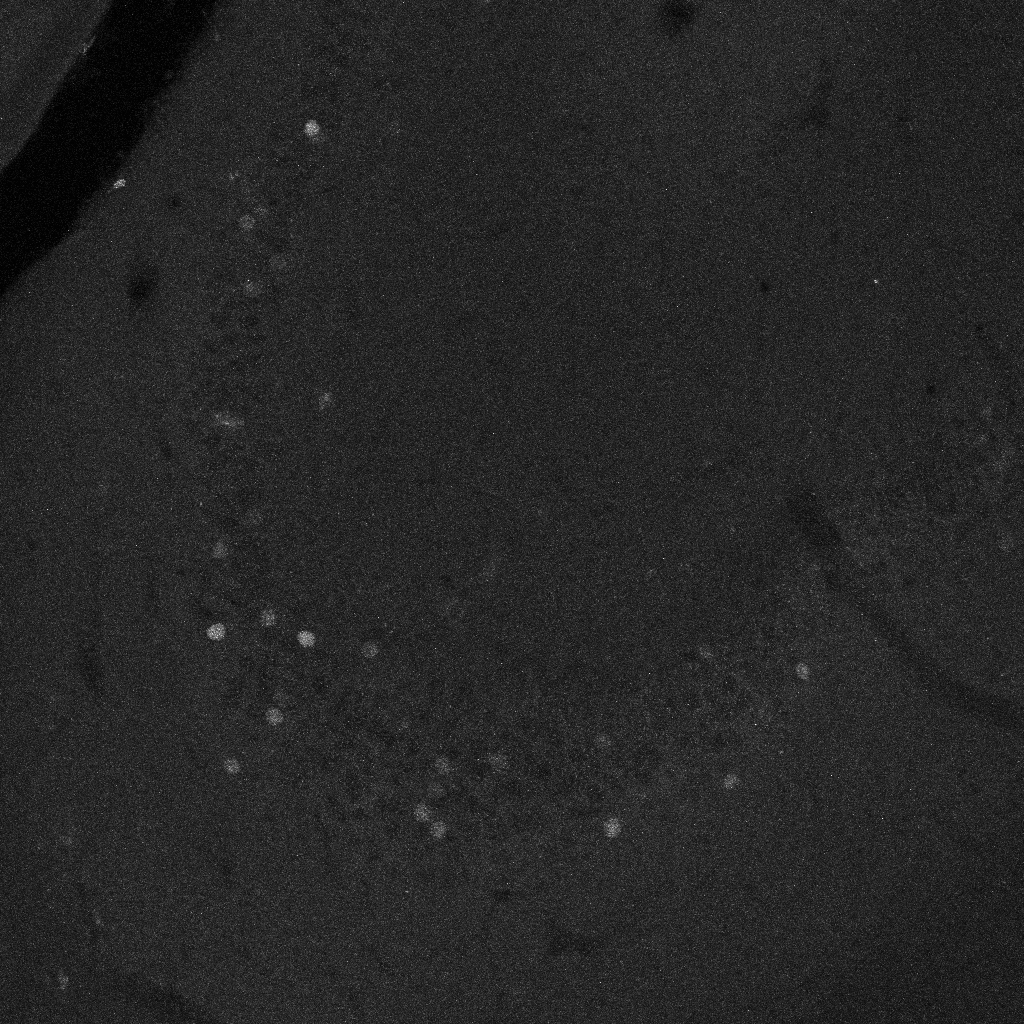

-
cFOS_Inns1: Fine-tuned on cFOS_Wue with five image-mask pairs of cFOS labels in the amygdala. Masks for fine-tuning were prepared by one expert. Images acquired using epifluorescence microscopy with a resolution of 1 pixel per µm.


-
cFOS_Inns2: Fine-tuned on cFOS_Wue with five image-mask pairs of cFOS labels in the infralimbic cortex. Masks for fine-tuning were prepared by one expert. Images acquired using epifluorescence microscopy with a resolution of 2 pixel per µm.

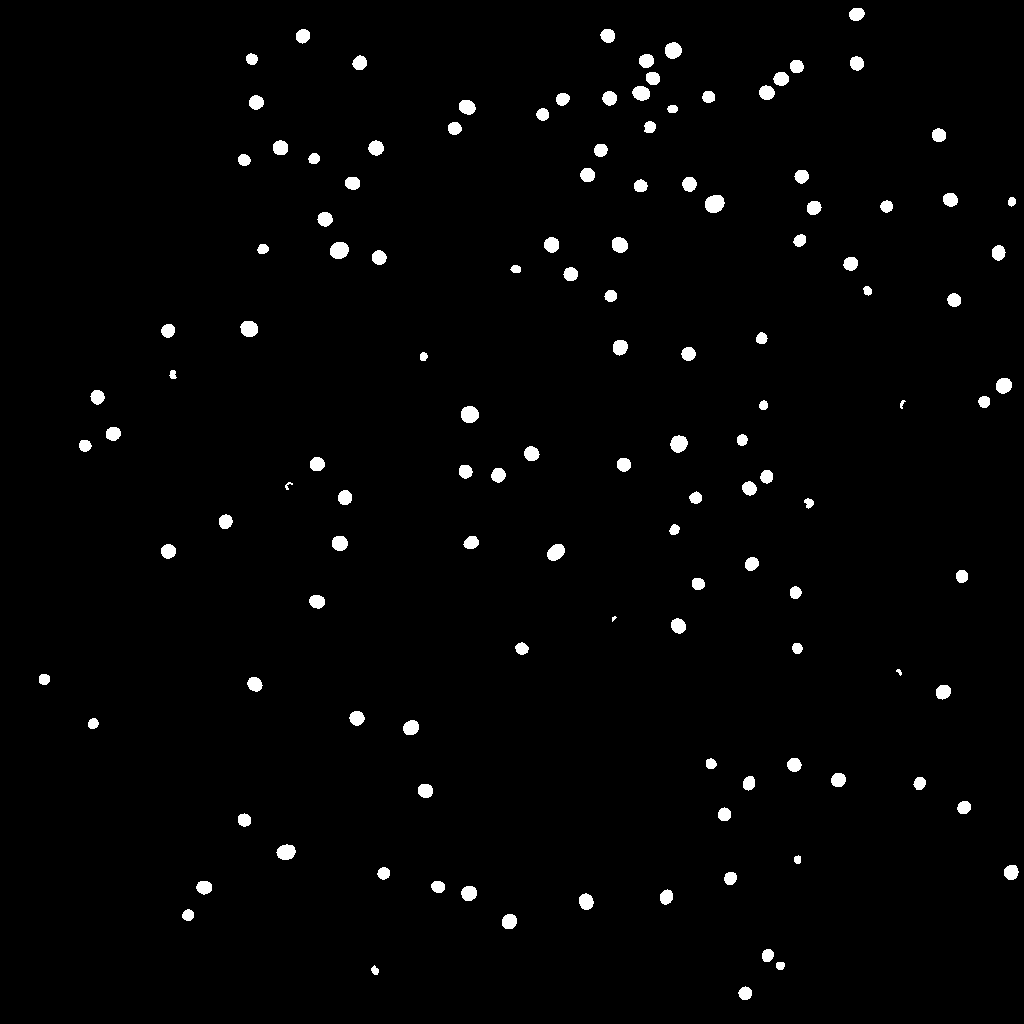
-
cFOS_Mue: Fine-tuned on cFOS_Wue with five image-mask pairs of cFOS labels in the paraventricular nucleus of the thalamus. Masks for fine-tuning were prepared by one expert. Images acquired using laser-scanning confocal microscopy with a resolution of 0.8 pixel per µm.


-
Parv: Trained on 36 image-mask pairs of Parvalbumin-labels in the dorsal hippocampus (including 12 images of each sub-region: dentate gyrus, CA3 and CA1). Masks for training were prepared by five independent experts. Images were acquired using laser-scanning confocal microscopy with a resolution of 1.6 pixel per µm.
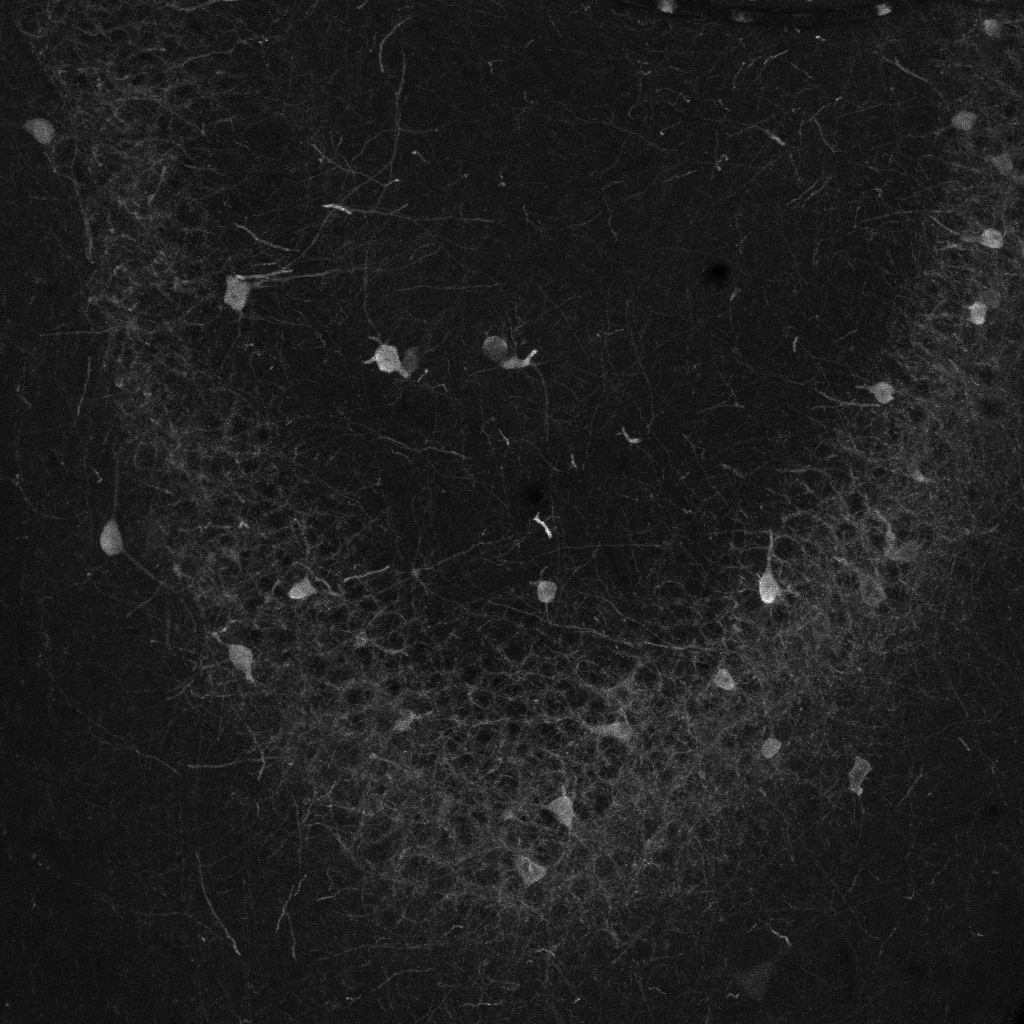

Creating segmentation maps for DeepFLaSH with Fiji/ImageJ
If you don't have labelled training data available, you can use this instruction manual for creating segmentation maps. The ImagJ-Macro is available here.
Citation
Use this bibtex to cite our paper:
@article{Segebarth2018,
author = {Segebarth, Dennis and Griebel, Matthias and Duerr, Alexander and R. von Collenberg, Cora and Martin, Corinna and Fiedler, Dominik and Comeras, Lucas B. and Sah, Anupam and Stein, Nikolai and Gupta, Rohini and Sasi, Manju and Lange, Maren D. and Tasan, Ramon O. and Singewald, Nicolas and Pape, Hans-Christian and Sendtner, Michael and Flath, Christoph M. and Blum, Robert},
title = {DeepFLaSh, a deep learning pipeline for segmentation of fluorescent labels in microscopy images},
year = {2018},
doi = {10.1101/473199},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2018/11/19/473199},
eprint = {https://www.biorxiv.org/content/early/2018/11/19/473199.full.pdf},
journal = {bioRxiv}
}
Acronym
A Deep-learning pipeline for Fluorescent Label Segmentation that learns from Human experts
