pallada-92 / Dna 3d Engine
Projects that are alternatives of or similar to Dna 3d Engine
cube3d.dna
The most advanced and compact 3d engine ever implemented in DNA code.






Getting started
How to deploy
- Synthesize the oligonucleotides from the cube3d.dna file.
- Arrange the test tubes as shown in the diagram below.
- Don't forget to provide the initial concentrations according to the table below.
- Use a pipette to encode the position (row and column) of each tube to start the computation.
Environment variables
q = 0.01
cxtm = 0.606
axp = 0.606
cytm = 0.898
ayp = 0.898
cztm = 1.243
azp = 1.243
mxyzm = 0.3
nx = 0.036 + 0.555 Col + 0.147 Row
ny = 0.853 + -0.517 Row
nz = 0.737 + -0.270 Col + 0.302 Row
Testing
- Pick the fluorophore of your favorite color and attach it to the
Strand R0, so that it activates when theRspecies are being produced. - Use a light source with a specific wavelength (depending on the fluorophore you've chosen) to render the result.
Disclaimer: perhaps it would not be a good idea to try this experiment in a real lab, because it will cost you a lot of money and most likely won’t work as intended the first time.
Ports to other languages
Gallery
 Ray marching common implementation 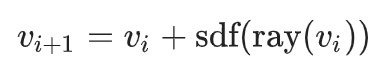
|
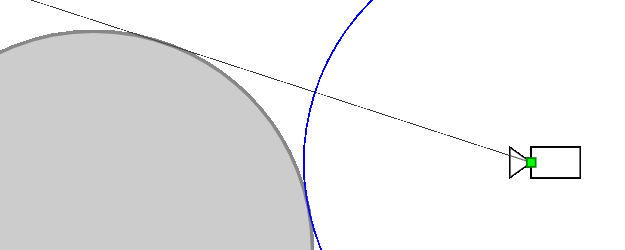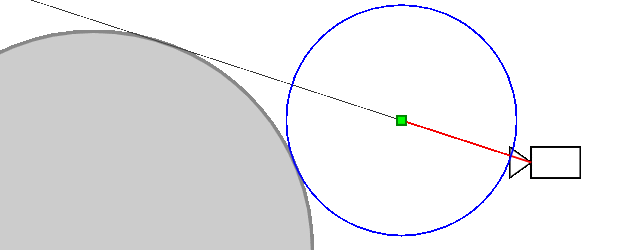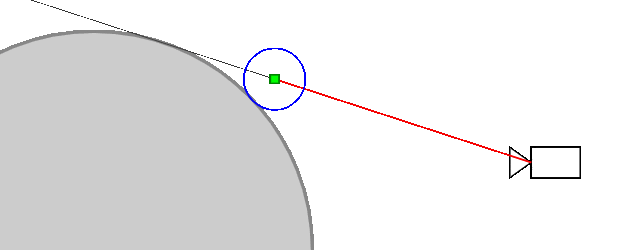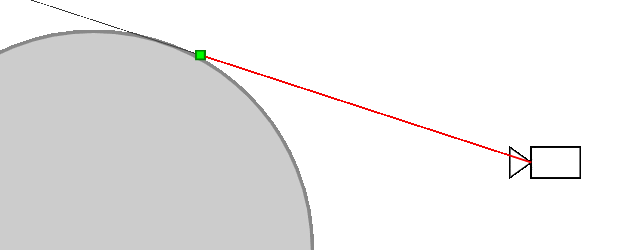 Ray marching differential form used here 
|
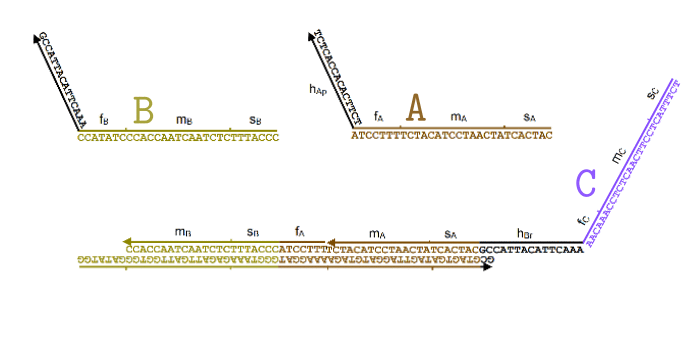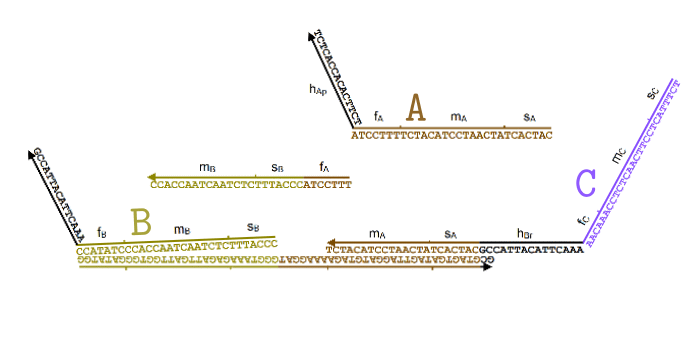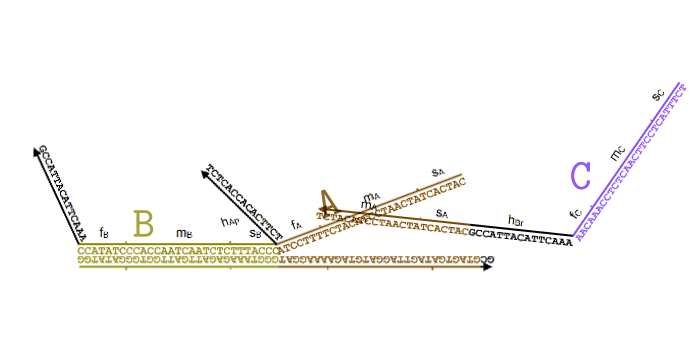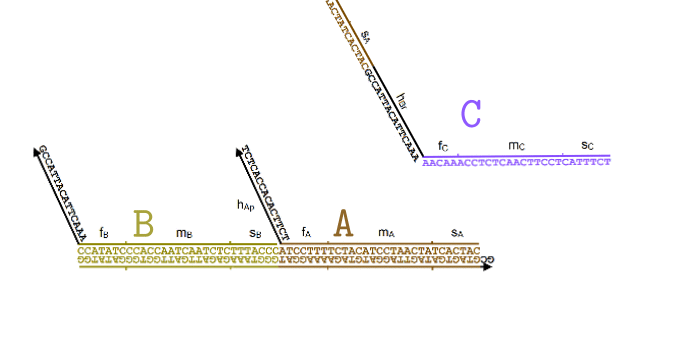 Simplified animation of the toehold mediated strand displacement technique (based on supplementary materials from [2]) |
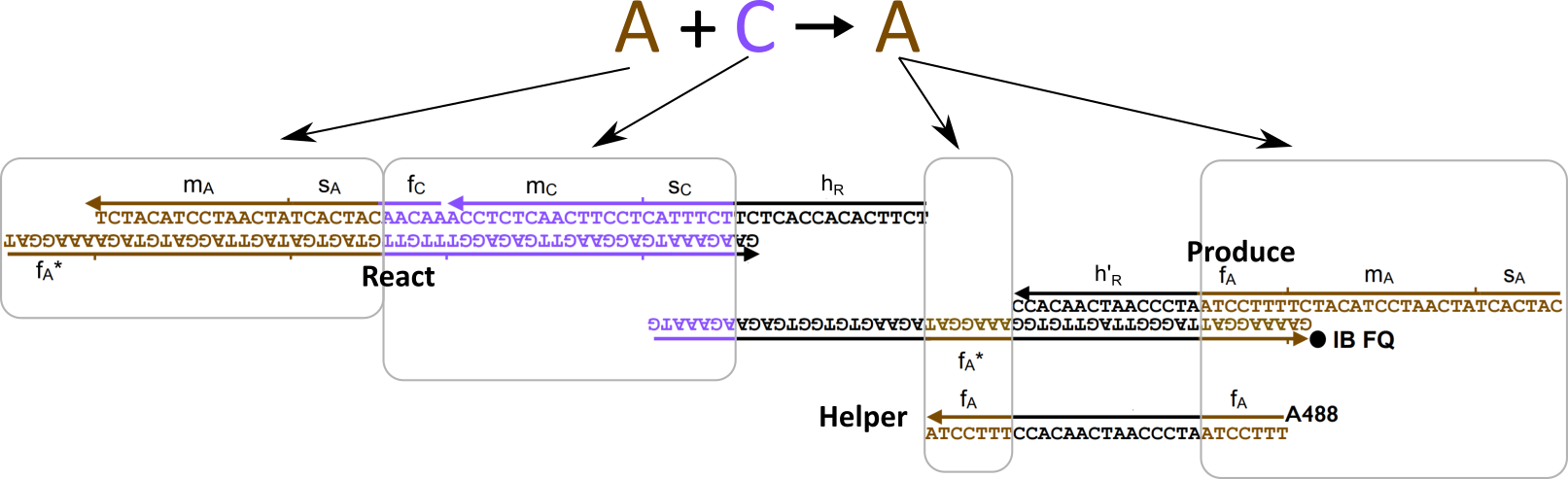 The types of oligonucleotides required for a single reaction (based on supplementary materials from [2]) |
 Data flow graph of the JS implementation. |
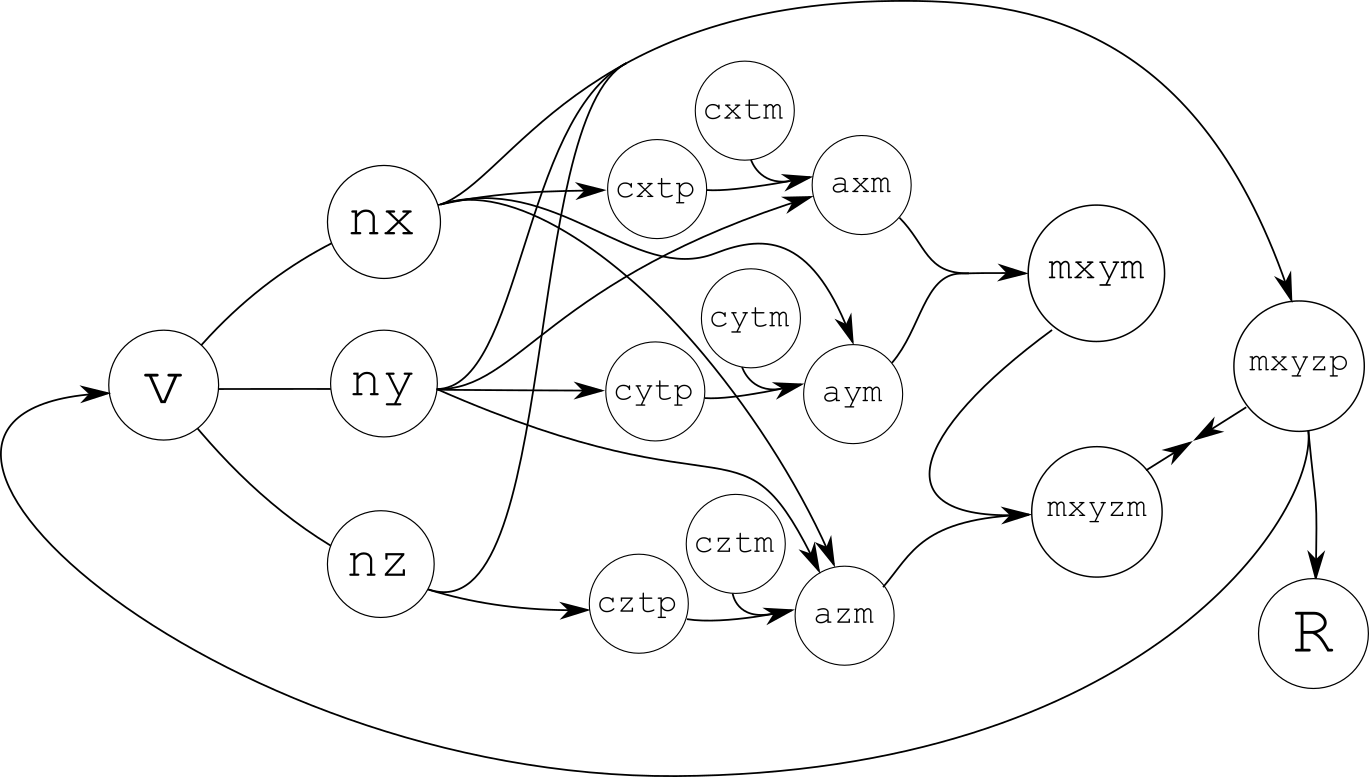 Species interaction graph of this implementation. |
 Unminified source code of this implementation which just comprises plain reactions with a few macros. The minifier was written in Wolfram Language specially for this project. |
 CRN++ source code for comparison (CRN++ is not used in this implementation). |
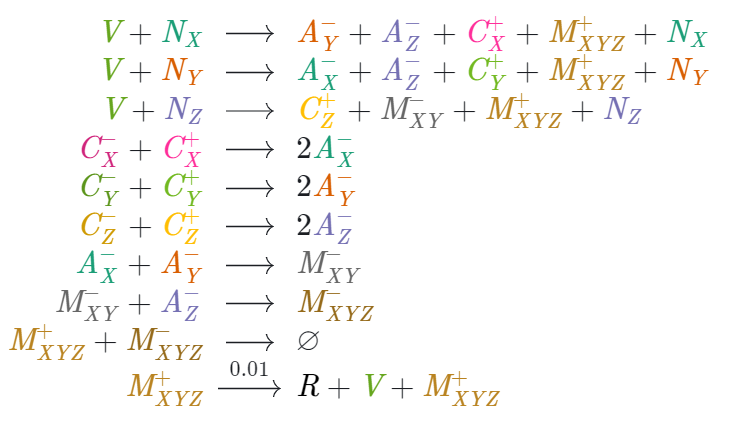 10 reactions after the minification used in this project. Since the piperine compiler doesn't support more than 2 products on the right-hand side, the reactions were split. |
 CRN++ 70 reactions output for comparison (CRN++ is not used in this implementation). |
Building from source
- Install the piperine compiler by the DNA and Natural Algorithms Group.
- Run the following command
piperine-design cube3d.crn --maxspurious 0.765
- Wait 2-3 hours for the compilation results.
References
-
David Soloveichik, Georg Seelig and Erik Winfree
DNA as a universal substrate for chemical kinetics
Proceedings of the National Academy of Sciences Mar 2010, 107 (12) 5393-5398; DOI: 10.1073/pnas.0909380107 -
Niranjan Srinivas, James Parkin, Georg Seelig, Erik Winfree and David Soloveichik
Enzyme-free nucleic acid dynamical systems
Science 358, eaal2052 (2017).
Some images were taken from supplementary materials -
Chalk, Cameron, Niels Kornerup, Wyatt Reeves, and David Soloveichik.
Composable rate-independent computation in continuous chemical reaction networks.
International Conference on Computational Methods in Systems Biology, pp. 256-273. Springer, Cham, 2018. -
Chen, Ho-Lin, David Doty, and David Soloveichik.
Rate-independent computation in continuous chemical reaction networks.
Proceedings of the 5th conference on Innovations in theoretical computer science. 2014. -
Marko Vasic, David Soloveichik and Sarfraz Khurshid
CRN++: Molecular Programming Language
Natural Computing (2020) 19:391–407 DOI: 10.1007/s11047-019-09775-1
CRN++ on GitHub -
Inigo Quilez
https://www.iquilezles.org/www/index.htm
Introduction to Raymarching Signed Distance Functions



