tevisgehr / Eeg Classification
Labels
Projects that are alternatives of or similar to Eeg Classification
EEG-Classification
This project is a joint effort with neurology labs at UNL and UCD Anschutz to use deep learning to classify EEG data.
The goal is to use various data processing techniques and deep neural network architectures to perserve both spacial and time information in the classification of EEG data.
For a more concise and visually pleasing presentation of this project, please see the included PDF. (galvanize_36x48_Tevis_Gehr_EEG_2.pdf)
Introduction
An electroencephalogram (EEG) is a test that detects electrical activity in your brain using small, flat metal discs (electrodes) attached to your scalp. Your brain cells communicate via electrical impulses and are active all the time, even when you're asleep. This activity shows up as wavy lines on an EEG recording. [Mayo Clinic]
The goal of this project is to classify brain states from EEG data. A joint CU Anschutz/ULN project has collected EEG data on subjects during sessions in which the subjects were instructed to visualize performing a motor-based task. Each subject performed one session visualizing a very familiar task, and another session visualizing an unfamiliar task. The primary goal is to develop a classifier that can correctly identify whether a subject is visualizing a task that is familiar or unfamiliar.
Secondary goals include providing insight into which brain regions and frequency bands associate with each of the respective classes. If a deep learning approach is found to be viable, these insights may correspond to latent features found within the neural network. Other insights may be obtained from more traditional data processing and machine learning techniques.
The Data
The data are in the form of csv files with raw waveform signals from 14 probes places around the scalp. The sampling rate is 128 hz, which allows for frequency analysis up to ~60 hz. Each of 8 subjects participated in two 1 minute sessions. Therefore the total number of datapoints is on the order of 14x128x60x8x2 = 1,720,320. Several additional subjects are expected to perform recording sessions during the next few weeks.
The image below shows the raw waveform data from four of the 14 channels during a typical session. EMG signals (such as those causes by swallowing or yawning) were manually removed.
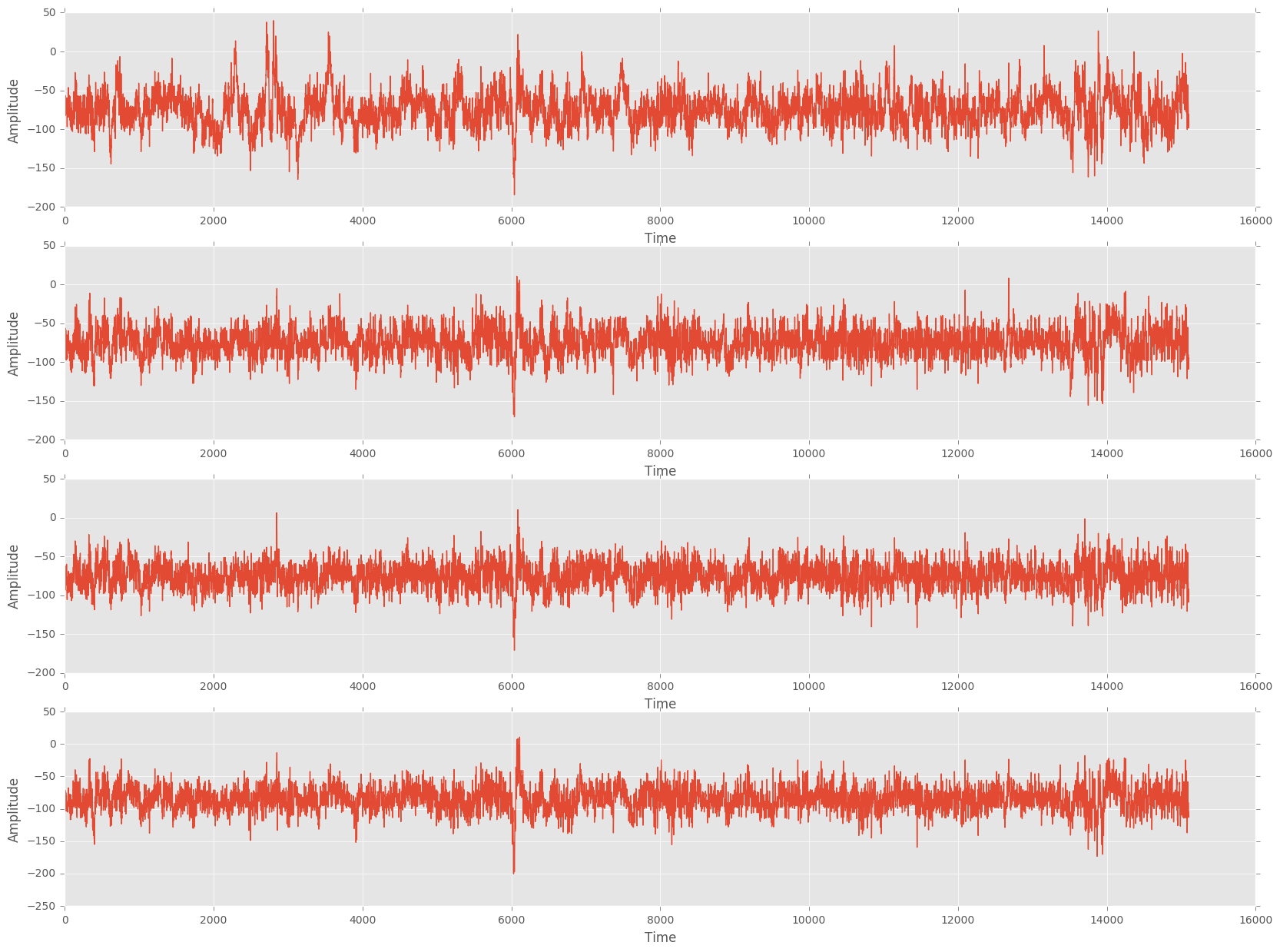
Figure 1: Raw waveform data from four of the 14 EEG probes
Tiers
The minimum result required for this project to be a full success is to have developed a classifier that is capable of accurately classifying snippets of EEG session data as being from the visualization of either a familiar or an unfamiliar skill. Because this is a binary classification problem with balanced classes, the minimum baseline for accuracy is 0.5. Full success would mean having an accuracy of at least 70% (although this number is arbitrary). State-of-the-art EEG classification techniques currently score considerably higher than this [1][2]. Data processing and augmentation is expected to be important and multiple approaches will be considered.
Once a viable classifier has been developed, the goals are twofold. First to use modern deep learning techniques to maximize the test accuracy. One proposed approach is outlined in a paper that proposes projecting the transformed EEG frequencies into a 2D image with a depth dimension for frequency band [1]. This format makes for ideal inputs into a standard convolutional neural network. This or another deep learning approach may be used to achieve the highest possible accuracy.
The second set of goals center around providing insight into the underlying mechanisms in brain function. The methods for accomplishment of these goals will depend on the details of the machine learning algorithms that are able to successfully classify the EEG data.
Approach
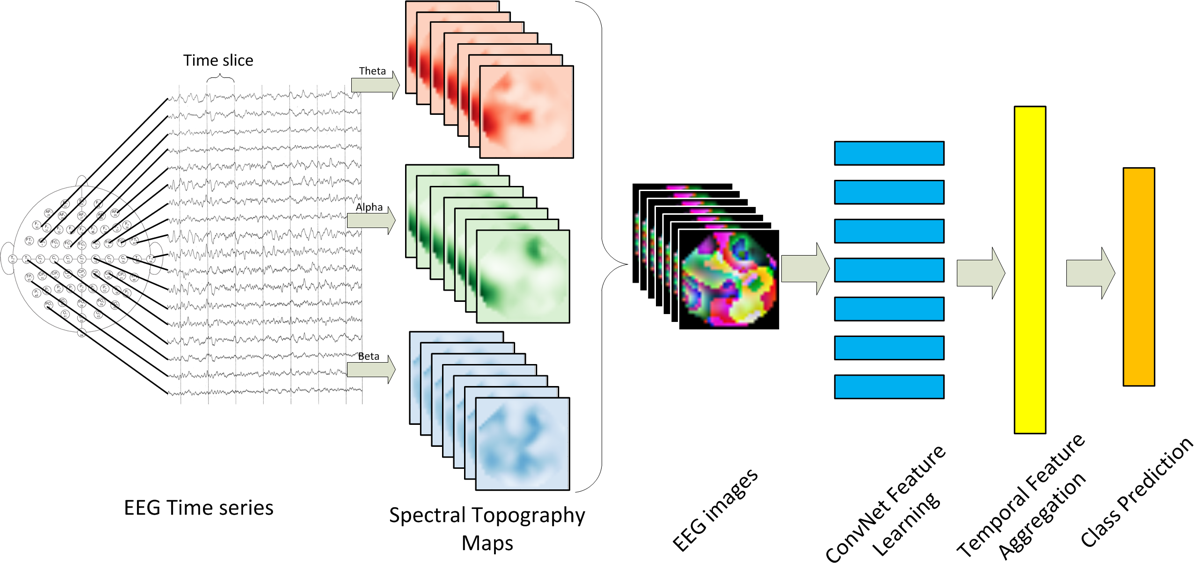
Relying on previous EEG research done by Beshivan et. al.[1], as well as the latest advances in video classification[3], the approach was to process the 14-channel time-series data into discreet one-second ‘frames’ and project these frames onto a 2D map of the surface of the head. Then a convolutional neural network (CNN) was trained to classify frames.
Figure 2:EEG classification architecture proposed by [1].
Data Processing
Following is a desciption of data processing techniques used in this project.
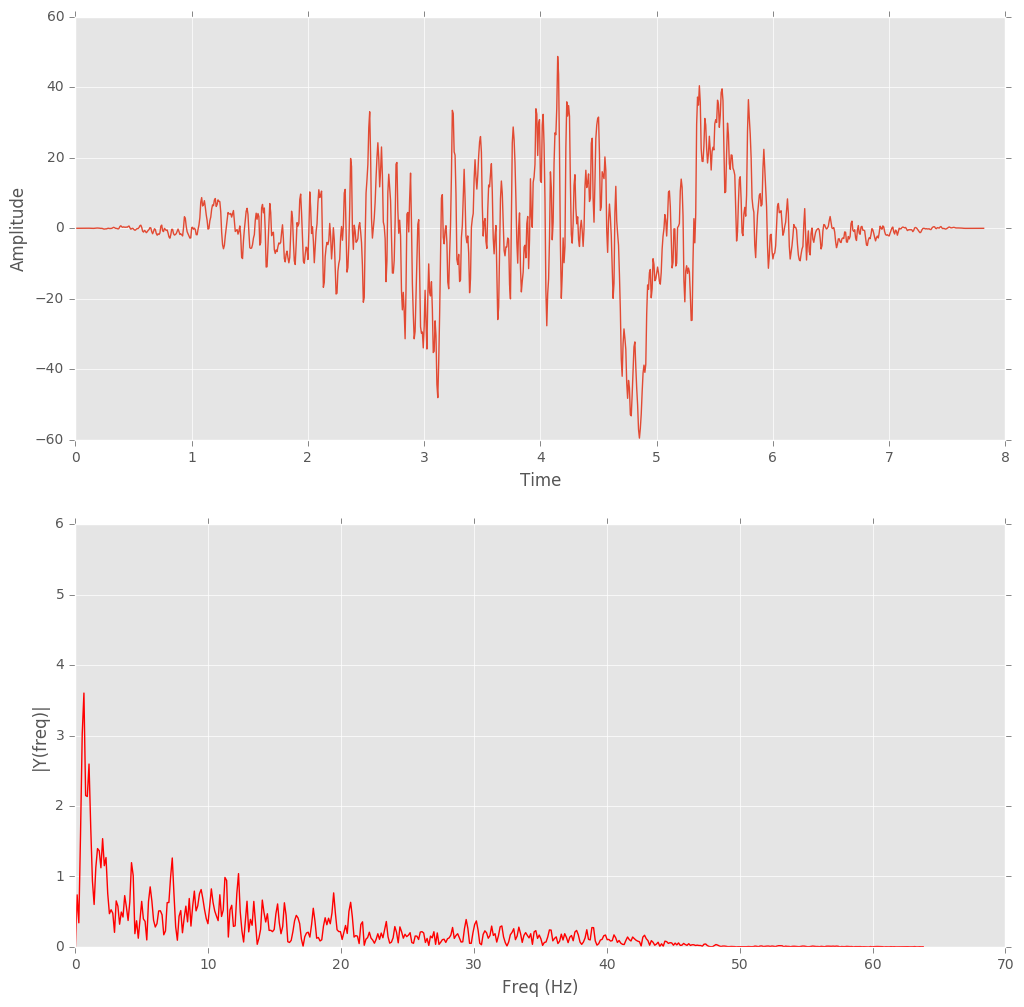
Hanning Window: First the data were chopped up into overlapping 1-second ‘frames’ and a Hanning window was applied.
Fast Fourier Transform(FFT): FFT was applied to transform data for each frame from time domain to frequency domain.
Frequency Binning: FFT amplitudes were grouped into theta(4-8Hz), alpha(8-12Hz), and beta(12-40Hz) ranges, giving 3 scalar values for each probe per frame.
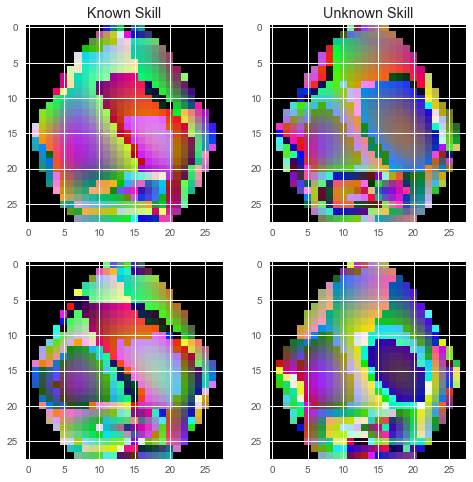
2D Azimuthal Projection: These 3 values were interpreted as RGB color channels and projected onto a 2D map of the head.
Figure 3: Hanning windowed one-second frame and FFT.
Overlapping one-second 'frames'.
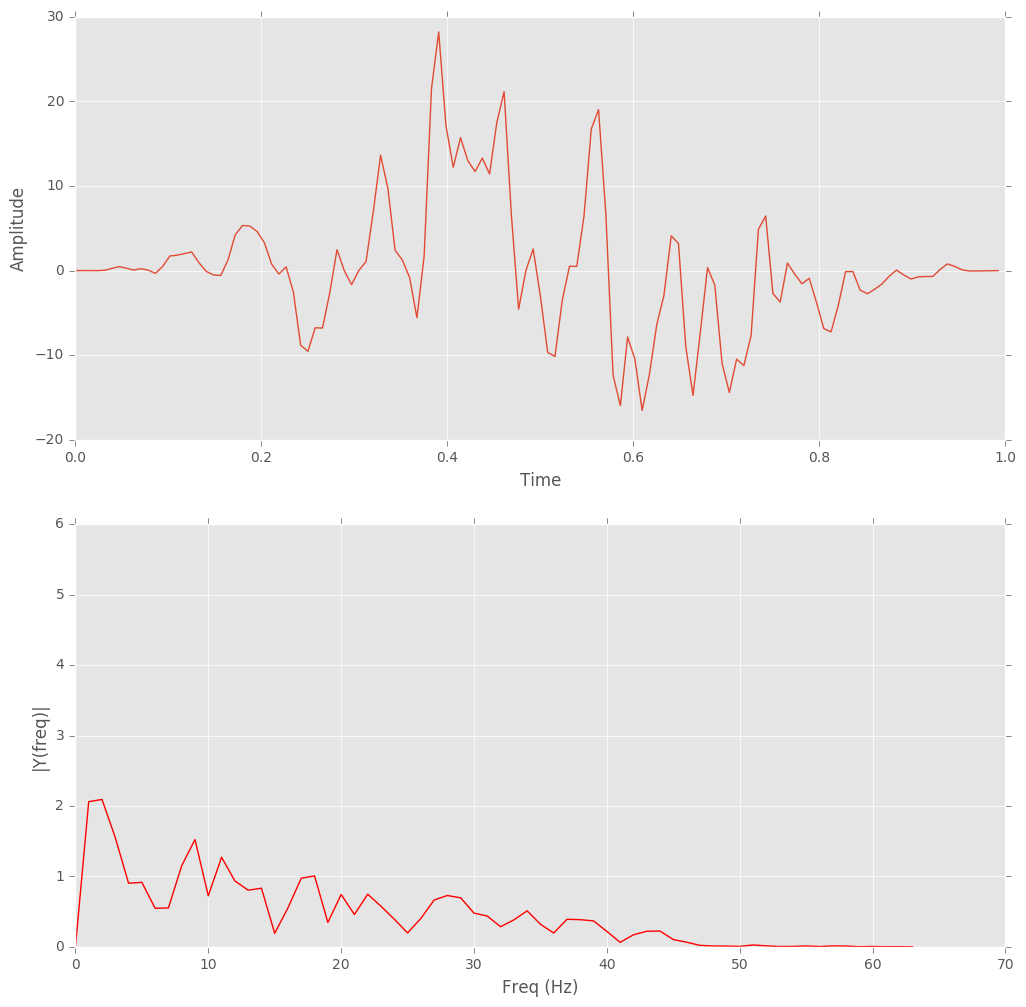
Figure 4: One 'frame'.
Figure 5: 2D projections of theta, alpha and beta ranges.
Network Architecture
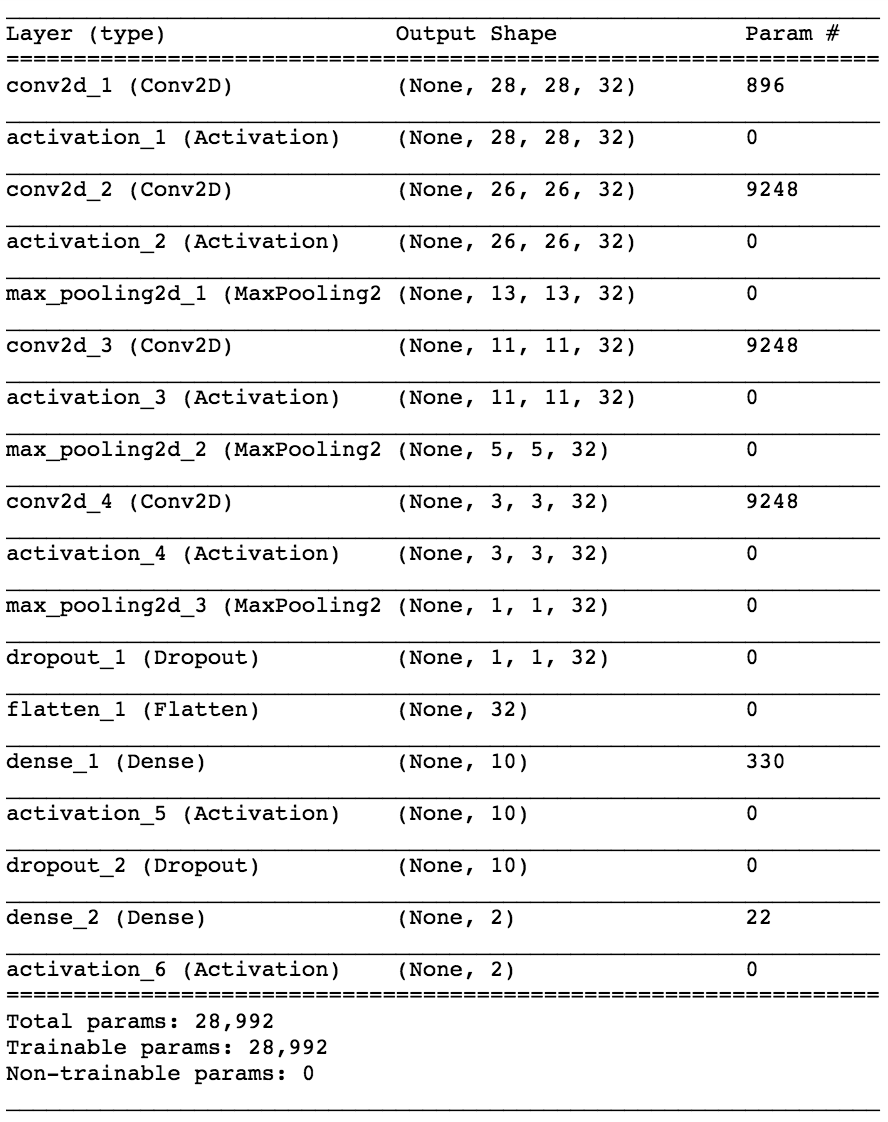
A convolutional neural network was iteratively constructed and tuned to give the best classification accuracy with the data availible. The final architecture is shown below.
Table 1: Summary of Convolutional Neural Network in Keras
Results and Discussion
The results obtained are encouraging. Without even using a recurrent neural network (which is the next logical step, see [1]), the CNN is able to correctly classify the test subject’s brain-state about 8.5 times out of 10. This is likely high enough to enable a new level of performance with brain-computer interface (BCI) technologies.
However, the best results were obtained when the network was trained on samples from the same recording session. While this may be practical for basic brain research, it would be less practical for use in BCI technology.
The results obtained suggest that while EEG signals do indeed generalize between individuals, there are still significant variations between individuals, which is an unsurprising finding.
This further suggests that using EEG for BCI will likely require an iterative approach of training on a large population and then fine tuning on a specific individual. It is therefore recommended that future research be done on the possible application of Transfer Learning techniques to the classification of EEG signals.
Citations
[1] Learning Representations from EEG with Deep Recurrent-Convolutional Neural Networks
19 Nov 2015. Bashivan et al. Cornell University Library.
https://arxiv.org/abs/1511.06448
[2] A novel deep learning approach for classification of EEG motor imagery signals
30 Nov 2016. Tabar and Halici. IOP Publishing.
http://iopscience.iop.org/article/10.1088/1741-2560/14/1/016003/meta
[3] Beyond Short Snippets: Deep Networks for Video Classification
13 Apr 2015. Ng et al. Cornell University Library.
https://arxiv.org/abs/1503.08909