IBCNServices / Gendis
Projects that are alternatives of or similar to Gendis
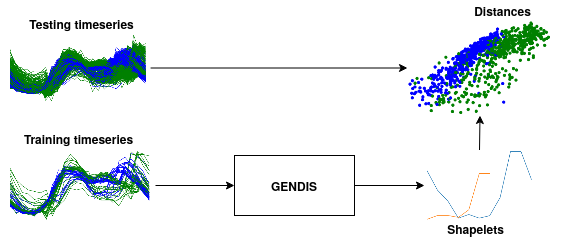
GENDIS 



GENetic DIscovery of Shapelets
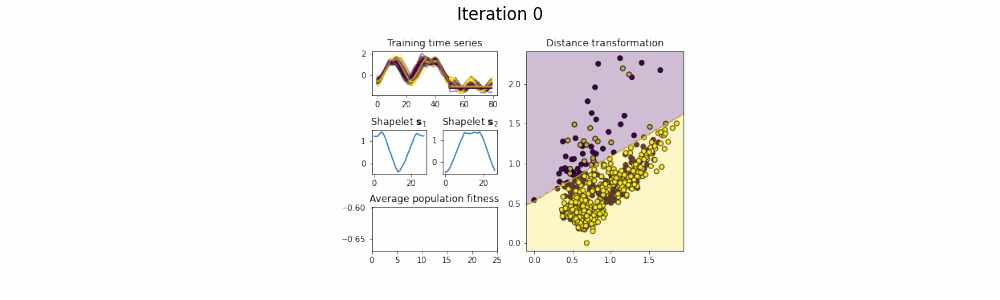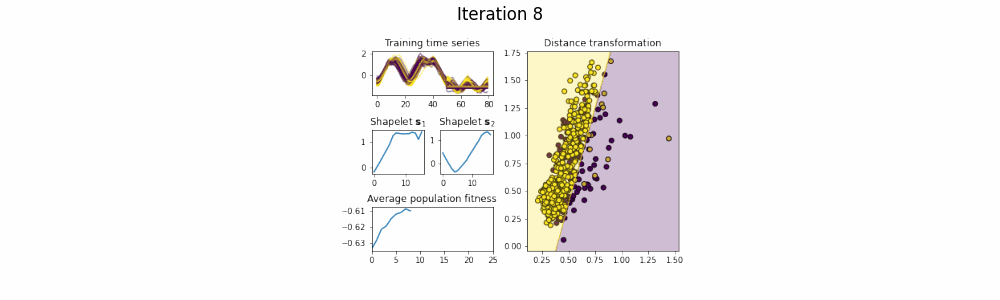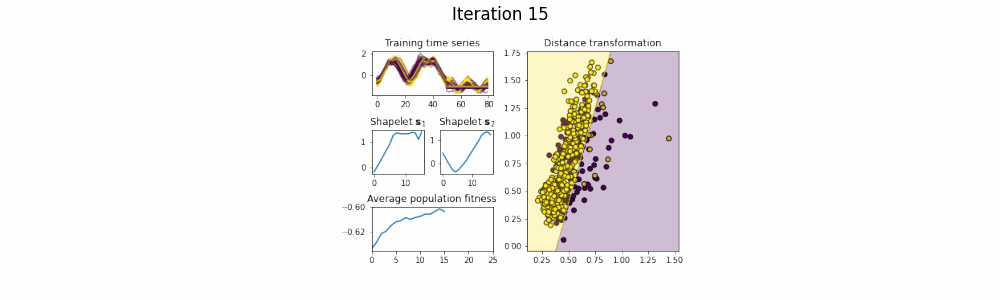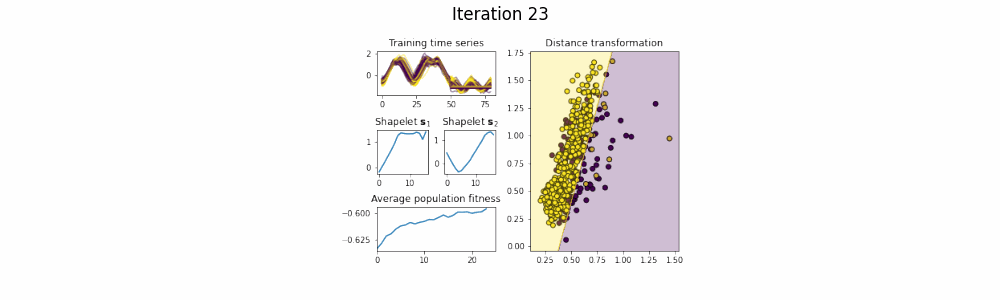
In the time series classification domain, shapelets are small subseries that are discriminative for a certain class. It has been shown that by projecting the original dataset to a distance space, where each axis corresponds to the distance to a certain shapelet, classifiers are able to achieve state-of-the-art results on a plethora of datasets.
This repository contains an implementation of GENDIS, an algorithm that searches for a set of shapelets in a genetic fashion. The algorithm is insensitive to its parameters (such as population size, crossover and mutation probability, ...) and can quickly extract a small set of shapelets that is able to achieve predictive performances similar (or better) to that of other shapelet techniques.
Installation
We currently support Python 3.5 & Python 3.6. For installation, there are two alternatives:
- Clone the repository
https://github.com/IBCNServices/GENDIS.gitand run(python3 -m) pip -r install requirements.txt - GENDIS is hosted on PyPi. You can just run
(python3 -m) pip install gendisto add gendis to your dist-packages (you can use it from everywhere).
Make sure NumPy and Cython is already installed (pip install numpy and pip install Cython), since that is required for the setup script.
Tutorial & Example
1. Loading & preprocessing the datasets
In a first step, we need to construct at least a matrix with timeseries (X_train) and a vector with labels (y_train). Additionally, test data can be loaded as well in order to evaluate the pipeline in the end.
import pandas as pd
# Read in the datafiles
train_df = pd.read_csv(<DATA_FILE>)
test_df = pd.read_csv(<DATA_FILE>)
# Split into feature matrices and label vectors
X_train = train_df.drop('target', axis=1)
y_train = train_df['target']
X_test = test_df.drop('target', axis=1)
y_test = test_df['target']
2. Creating a GeneticExtractor object
Construct the object. For a list of all possible parameters, and a description, please refer to the documentation in the code
from gendis.genetic import GeneticExtractor
genetic_extractor = GeneticExtractor(population_size=50, iterations=25, verbose=True,
mutation_prob=0.3, crossover_prob=0.3,
wait=10, max_len=len(X_train) // 2)
3. Fit the GeneticExtractor and construct distance matrix
shapelets = genetic_extractor.fit(X_train, y_train)
distances_train = genetic_extractor.transform(X_train)
distances_test = genetic_extractor.transform(X_test)
4. Fit ML classifier on constructed distance matrix
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
lr = LogisticRegression()
lr.fit(distances_train, y_train)
print('Accuracy = {}'.format(accuracy_score(y_test, lr.predict(distances_test))))
Example notebook
A simple example is provided in this notebook
Data
All datasets in this repository are downloaded from timeseriesclassification. Please refer to them appropriately when using any dataset.
Paper experiments
In order to reproduce the results from the corresponding paper, please check out this directory.
Tests
We provide a few doctests and unit tests. To run the doctests: python3 -m doctest -v <FILE>, where <FILE> is the Python file you want to run the doctests from. To run unit tests: nose2 -v
Contributing, Citing and Contact
If you have any questions, are experiencing bugs in the GENDIS implementation, or would like to contribute, please feel free to create an issue/pull request in this repository or take contact with me at gilles(dot)vandewiele(at)ugent(dot)be
If you use GENDIS in your work, please use the following citation:
@article{vandewiele2021gendis,
title={GENDIS: Genetic Discovery of Shapelets},
author={Vandewiele, Gilles and Ongenae, Femke and Turck, Filip De},
journal={Sensors},
volume={21},
number={4},
pages={1059},
year={2021},
publisher={Multidisciplinary Digital Publishing Institute}
}