brain-research / Guided Evolutionary Strategies
Labels
Projects that are alternatives of or similar to Guided Evolutionary Strategies
Guided Evolutionary Strategies
Link to demo notebooks:
Link to paper: arXiv/1806.10230
Overview
Many applications in machine learning require optimizing a function whose true gradient is unknown, but where surrogate gradient information (directions that may be correlated with, but not necessarily identical to, the true gradient) is available instead. This arises when an approximate gradient is easier to compute than the full gradient (e.g. in meta-learning or unrolled optimization), or when a true gradient is intractable and is replaced with a surrogate (e.g. in certain reinforcement learning applications, or when using synthetic gradients).
Here, we propose Guided Evolutionary Strategies (Guided ES), a method for optimally using surrogate gradient directions along with random search. We define a search distribution for evolutionary strategies that is elongated along a guiding subspace spanned by the surrogate gradients. This allows us to estimate a descent direction which can then be passed to a first-order optimizer.
This repository contains a colaboratory (colab) notebook with a demo of the method on a toy problem (described below).
Introduction
Imagine you have a function you would like to optimize, but you only have access to approximate gradients of the function. There are two approaches to optimization. On one hand, you could ignore the surrogate gradient information entirely and perform zeroth-order optimization, using methods such as evolutionary strategies to estimate a descent direction. These methods exhibit poor convergence properties when the parameter dimension is large. On the other hand, you could directly feed the surrogate gradients to a first-order optimization algorithm. However, bias in the surrogate gradients will interfere with optimizing the target problem. Ideally, we would like a method that combines the complementary strengths of these two approaches: we would like to combine the unbiased descent direction estimated with evolutionary strategies with the low-variance estimate given by the surrogate gradient. We propose a method for doing this called guided evolutionary strategies (Guided ES).
Method
Our idea is to keep track of a low-dimensional subspace defined by the recent history of surrogate gradients during optimization (inspired by quasi-Newton methods) which we call the guiding subspace.
We then perform a finite difference random search (as in evolutionary strategies) preferentially within this subspace. By concentrating our search samples in a low-dimensional subspace where the true gradient has non-negligible support, we can dramatically reduce the variance of our search direction.
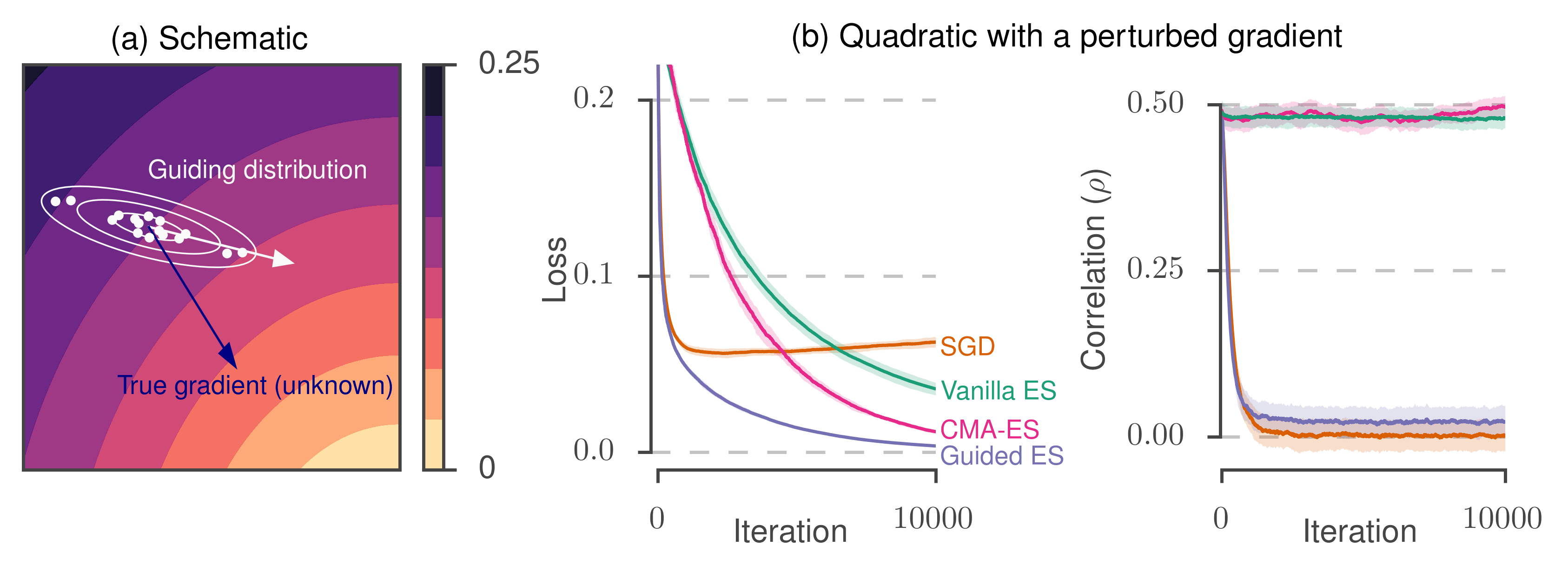
The figure panel (a) below depicts the geometry underlying our method. Instead of the true gradient (blue arrow), we are given a surrogate gradient (white arrow) which is correlated with the true gradient. We use this to form a guiding distribution (denoted with white contours) and use this to draw samples (white dots) which we use as part of a random search procedure.
In panel (b), we demonstrate the performance of the method on a toy problem. The problem consists of a random quadratic function, where we add an explicit bias and random noise to the gradient. Following the gradient directly with SGD (orange curve) starts fast but starts to diverge due to the bias in the gradient. Performing evolutionary strategies (or an adaptive variant, CMA-ES) succeed in minimizing the true function but proceed slowly and ignore the gradient information.
Guided ES, on the other hand, combines the strengths of these two approaches.
Citation
If you use this code, please consider citing our paper:
@article{
maheswaranathan2018guided,
title = {Guided evolutionary strategies: escaping the curse of dimensionality in random search},
author = {Niru Maheswaranathan and Luke Metz and Dami Choi and George Tucker and Jascha Sohl-Dickstein},
year = {2018},
eprint = {arXiv:1806.10230},
url = {https://arxiv.org/abs/1806.10230},
}
Contact
Authors:
- Niru Maheswaranathan ([email protected])
- Luke Metz ([email protected])
- Dami Choi ([email protected])
- George Tucker ([email protected])
- Jascha Sohl-Dickstein ([email protected])
This is not an officially supported Google product.