junqiangchen / Kits19 Challege
KiTS19——2019 Kidney Tumor Segmentation Challenge
Stars: ✭ 91
Programming Languages
python
139335 projects - #7 most used programming language
Labels
Projects that are alternatives of or similar to Kits19 Challege
Deep Segmentation
CNNs for semantic segmentation using Keras library
Stars: ✭ 69 (-24.18%)
Mutual labels: segmentation
Rgbd semantic segmentation pytorch
PyTorch Implementation of some RGBD Semantic Segmentation models.
Stars: ✭ 84 (-7.69%)
Mutual labels: segmentation
Mit Deep Learning
Tutorials, assignments, and competitions for MIT Deep Learning related courses.
Stars: ✭ 8,912 (+9693.41%)
Mutual labels: segmentation
Fcn.tensorflow
Tensorflow implementation of Fully Convolutional Networks for Semantic Segmentation (http://fcn.berkeleyvision.org)
Stars: ✭ 1,230 (+1251.65%)
Mutual labels: segmentation
Litiv
C++ implementation pool for computer vision R&D projects.
Stars: ✭ 82 (-9.89%)
Mutual labels: segmentation
Torch Points3d
Pytorch framework for doing deep learning on point clouds.
Stars: ✭ 1,135 (+1147.25%)
Mutual labels: segmentation
Niftynet
[unmaintained] An open-source convolutional neural networks platform for research in medical image analysis and image-guided therapy
Stars: ✭ 1,276 (+1302.2%)
Mutual labels: segmentation
Prin
Pointwise Rotation-Invariant Network (AAAI 2020)
Stars: ✭ 81 (-10.99%)
Mutual labels: segmentation
Caffe Model
Caffe models (including classification, detection and segmentation) and deploy files for famouse networks
Stars: ✭ 1,258 (+1282.42%)
Mutual labels: segmentation
Cnn Paper2
🎨 🎨 深度学习 卷积神经网络教程 :图像识别,目标检测,语义分割,实例分割,人脸识别,神经风格转换,GAN等🎨🎨 https://dataxujing.github.io/CNN-paper2/
Stars: ✭ 77 (-15.38%)
Mutual labels: segmentation
Pointclouddatasets
3D point cloud datasets in HDF5 format, containing uniformly sampled 2048 points per shape.
Stars: ✭ 80 (-12.09%)
Mutual labels: segmentation
Quicknat pytorch
PyTorch Implementation of QuickNAT and Bayesian QuickNAT, a fast brain MRI segmentation framework with segmentation Quality control using structure-wise uncertainty
Stars: ✭ 74 (-18.68%)
Mutual labels: segmentation
Brats17
Patch-based 3D U-Net for brain tumor segmentation
Stars: ✭ 85 (-6.59%)
Mutual labels: segmentation
Multiclass Semantic Segmentation Camvid
Tensorflow 2 implementation of complete pipeline for multiclass image semantic segmentation using UNet, SegNet and FCN32 architectures on Cambridge-driving Labeled Video Database (CamVid) dataset.
Stars: ✭ 67 (-26.37%)
Mutual labels: segmentation
Cws
Source code for an ACL2016 paper of Chinese word segmentation
Stars: ✭ 81 (-10.99%)
Mutual labels: segmentation
Ai Challenger Retinal Edema Segmentation
DeepSeg : 4th place(top3%) solution for Retinal Edema Segmentation Challenge in 2018 AI challenger
Stars: ✭ 88 (-3.3%)
Mutual labels: segmentation
Vnet Tensorflow
Tensorflow implementation of the V-Net architecture for medical imaging segmentation.
Stars: ✭ 84 (-7.69%)
Mutual labels: segmentation
KiTS19——2019 Kidney Tumor Segmentation Challenge
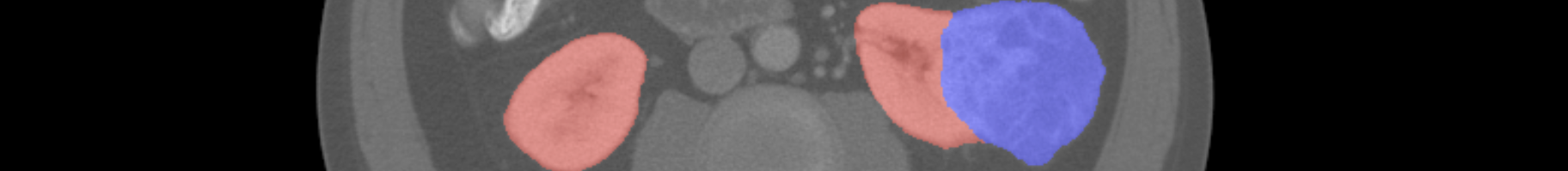
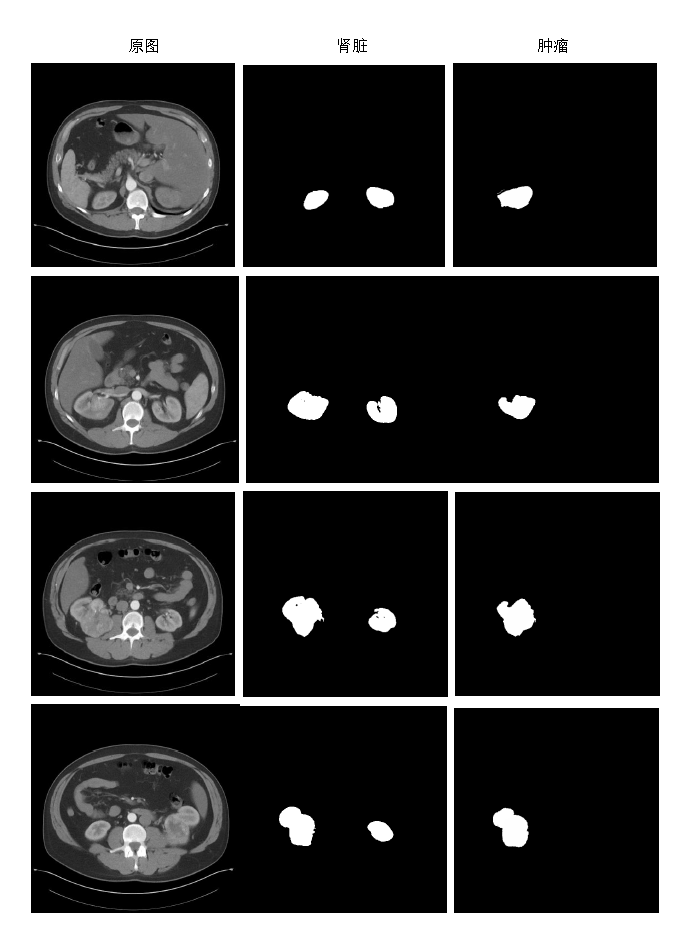
This is an example of the CT images Kidney Tumor Segmentation
Prerequisities
The following dependencies are needed:
- python == 3.5.5
- numpy >= 1.11.1
- SimpleITK >= 1.0.1
- opencv-python >= 3.3.0
- tensorflow-gpu == 1.8.0
- pandas >=0.20.1
- scikit-learn >= 0.17.1
- json >=2.0.9
How to Use
1、Preprocess
- analyze the ct image,and get the slice thickness and window width and position:run the dataAnaly.py
1.1 Preprocess Kidney
- keep kidney region into fixed size(512,512,64) for Corse Kidney Segmentation:run the corsedata2dprepare.py
- generate patch(128,128,64) kidney image and mask for Corse Kidney Segmentation:run the corsedata3dprepare.py
- keep Kidney region range image for fine Kidney Segmentation:run the finedata2dprepare.py
- generate patch(128,128,64) kidney image and mask for fine Kidney Segmentation:run the finedata3dprepare.py
- save patch image and mask into csv file: run the utils.py,like file trainSegmentation.csv
- split trainSegmentation.csv into training set and test set:run subset.py
1.2 Preprocess Kidney Tumor
- generate tumor image and mask for 2d Kidney Tumor Segmentation:run the tumordata2dprepare.py
- generate tumor image and mask for 3d Kidney Tumor Segmentation:run the finedata3dprepare.py
- save tumor image and mask path into csv file: run the utils.py,like file traintumorSegmentation.csv
- split traintumorSegmentation.csv into training set and test set
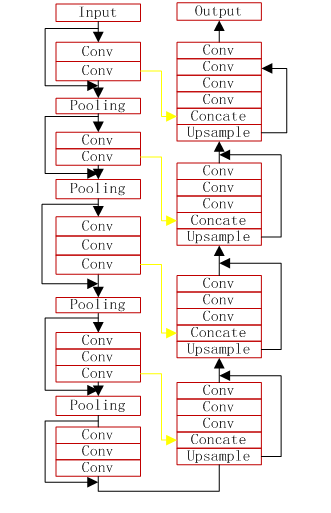
2、Kidney Segmentation
- the VNet model
2.1 Corse Kidney Segmentation
- Corse Kidney Segmentation training:run the train_vnet3d_kidney_corse.py
- Corse Kidney Segmentation inference:run the inference_vnet3d_kidney_corse.py
- this step get Corse Kidney range,can find the start and end pos in the kidneyrang.txt
2.2 Fine Kidney Segmentation
- Fine Kidney Segmentation training:run the train_vnet3d_kidney_fine.py
- Fine Kidney Segmentation inference:run the inference_vnet3d_kidney_fine.py
- this step following the 2.1 result,get fine Kidney result
2.3 Fine Kidney Segmentation
- remove Kidney Segmentation small object:run the segresultprocess.py removekidneysmallobj function
3、Kidney Tumor Segmentation
- the VNet2d model
3.1 2d Kidney Tumor Segmentation
-
2d Kidney Tumor Segmentation training:run the train_vnet2d_tumor.py
-
2d Kidney Tumor Segmentation inference:run the inference_vnet2d_tumor.py
-
this step get 2d slice tumor result
-
the VNet3d model
3.2 3d Kidney Tumor Segmentation
- 3d Kidney Tumor Segmentation training:run the train_vnet3d_tumor.py
- 3d Kidney Tumor Segmentation inference:run the inference_vnet3d_tumor.py
- this step get 3d tumor result
3.3 Kidney Tumor Result Process
- remove Kidney Tumor Segmentation small object:run the segresultprocess.py remove2d3dtumorsmallobj function
- calculate overlap between 2d tumor and 3d tumor reslut.
- save the region of 2d tumor and 3d tumor reslut that connect overlap region.
- save the region of 2d tumor and 3d tumor within Kidney result.
- merge the above two result and get final tumor result
- all above step can find in the segresultprocess.py
Result
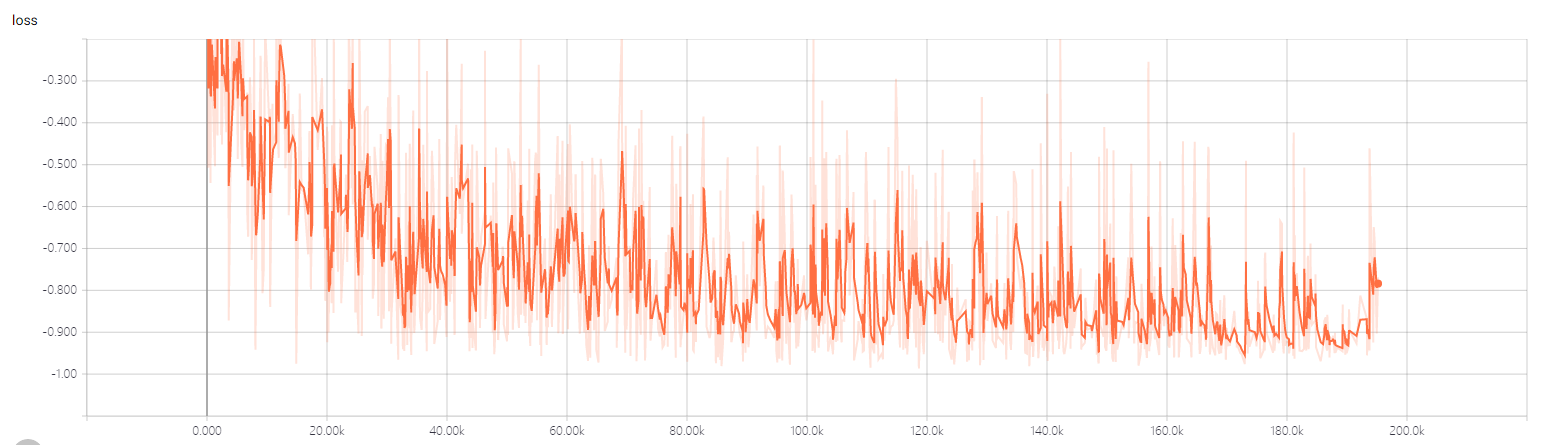
1、Kidney Segmentation
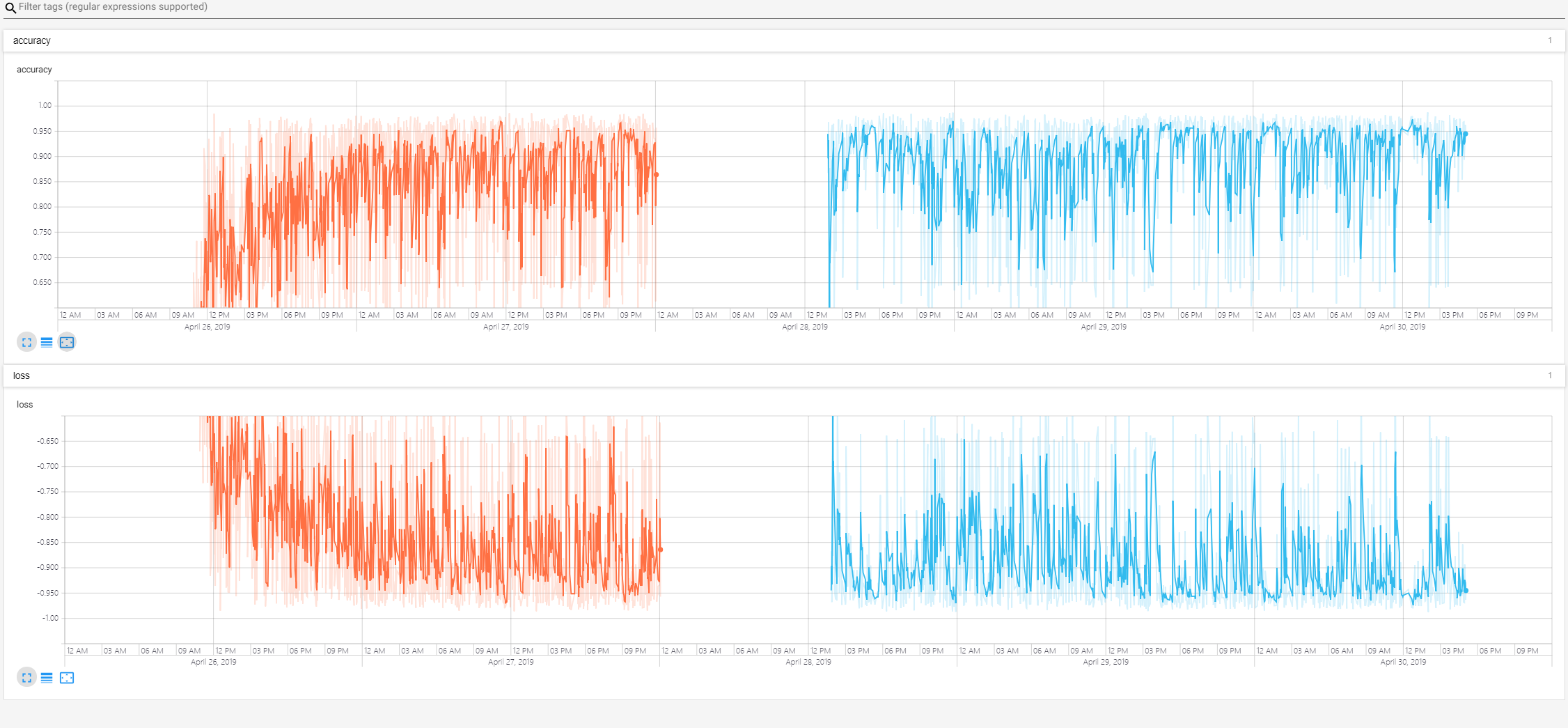
- the train loss
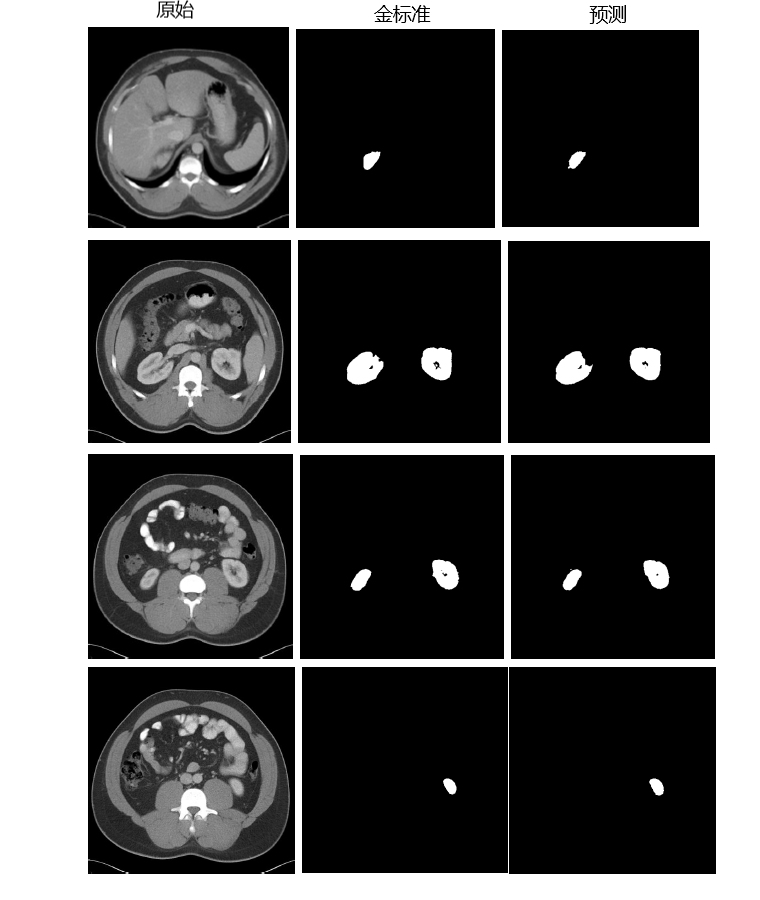
- 200-209case dice value and result
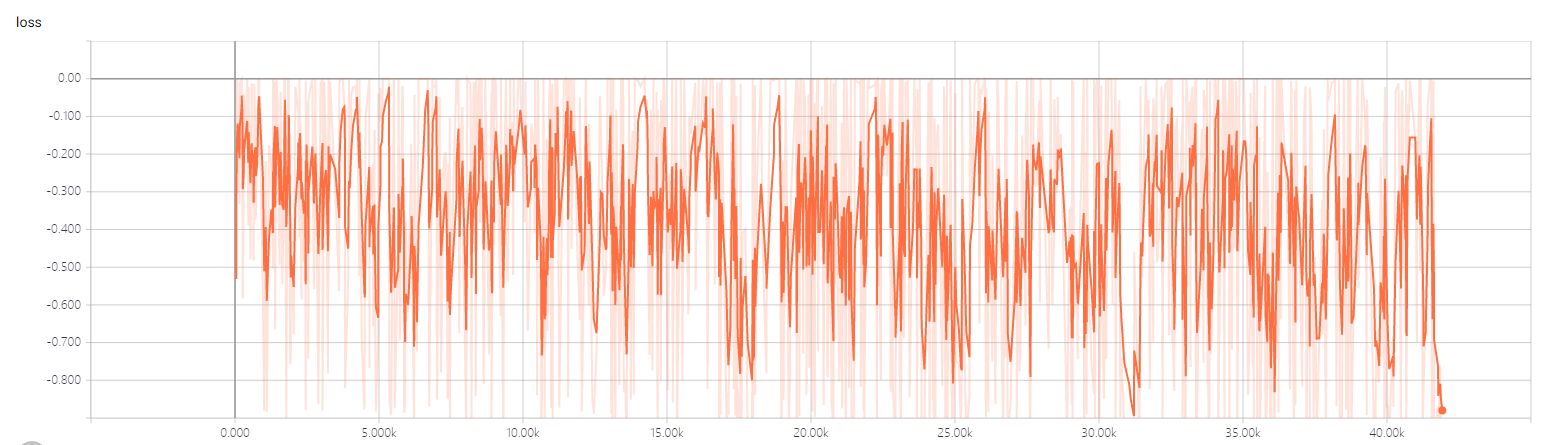
2、Kidney Tumor Segmentation
- the 2dtrain loss
- the 3dtrain loss
3、Lead Board
Contact
- https://github.com/junqiangchen
- email: [email protected],[email protected]
- Contact: junqiangChen,junMa(马骏)
- WeChat Number: 1207173174
- WeChat Public number: 最新医学影像技术
Note that the project description data, including the texts, logos, images, and/or trademarks,
for each open source project belongs to its rightful owner.
If you wish to add or remove any projects, please contact us at [email protected].