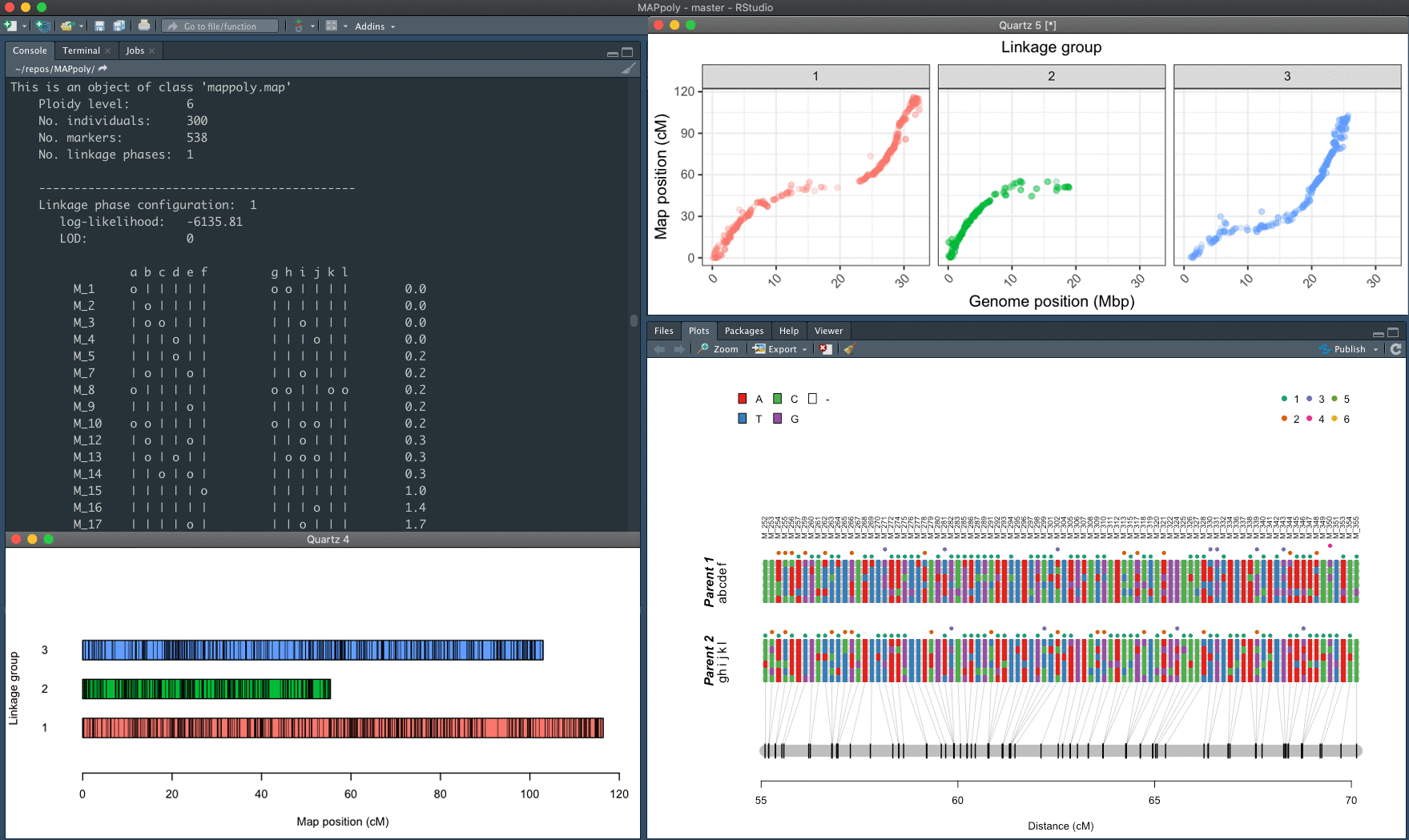
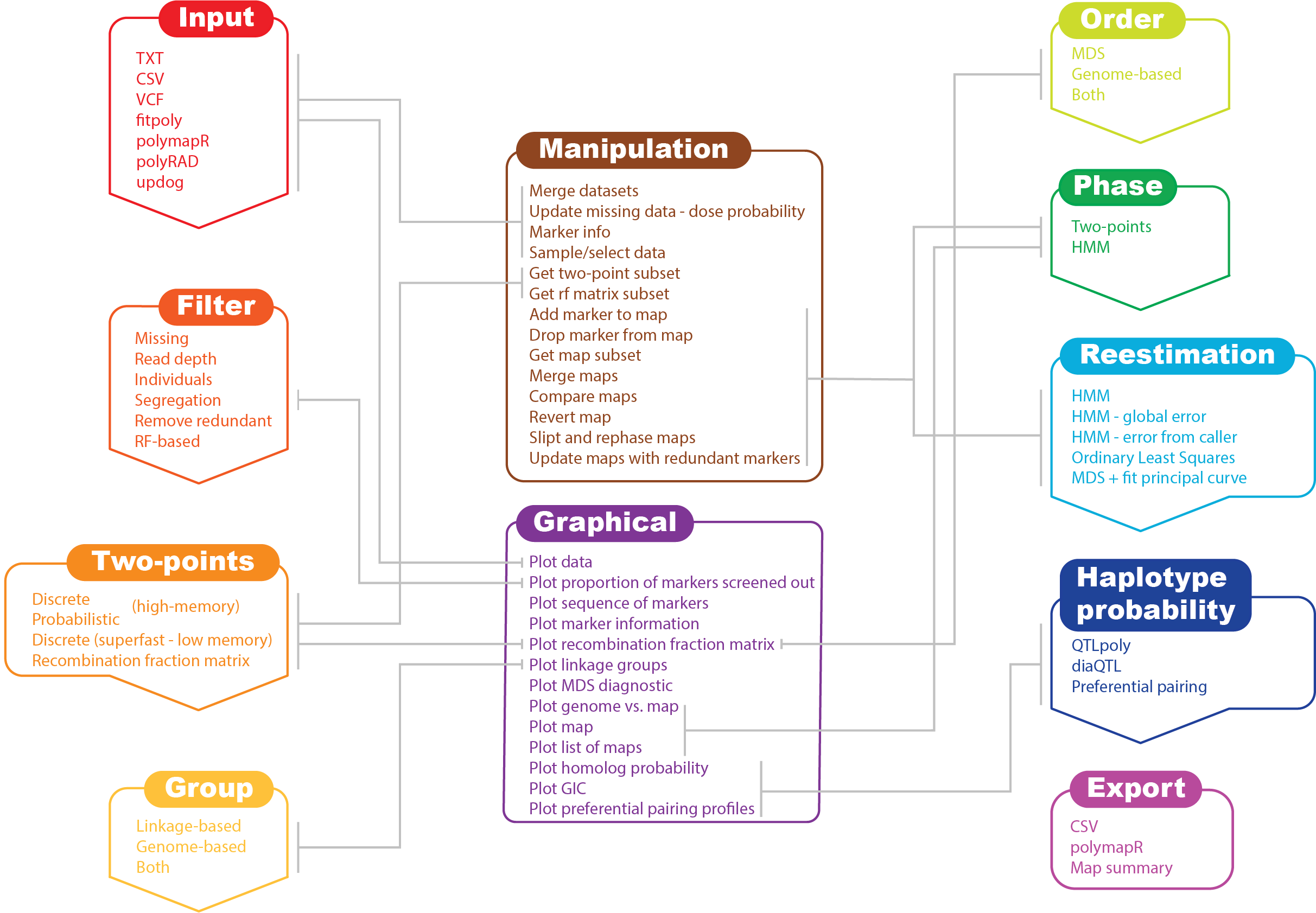
MAPpoly 
MAPpoly (v. 0.3.0) is an R package to construct genetic maps in autopolyploids with even ploidy levels. In its current version, MAPpoly can handle ploidy levels up to 8 when using hidden Markov models (HMM), and up to 12 when using the two-point simplification. When dealing with large numbers of markers (> 10,000), we strongly recommend using high-performance computation.
In its current version, MAPpoly can handle the following types of datasets:
- CSV files
- MAPpoly files
- Dosage based
- Probability based
- fitPoly files
- VCF files
MAPpoly also is capable of importing objects generated by the following R packages
The mapping strategy is based on using pairwise recombination fraction estimation as the first source of information to position allelic variants in specific homologues sequentially. For situations where pairwise analysis has limited power, the algorithm relies on the multilocus likelihood obtained through a hidden Markov model (HMM). The derivation of the HMM used in MAPpoly can be found in Mollinari and Garcia, 2019.
Installation
From CRAN (stable version)
To install MAPpoly from the The Comprehensive R Archive Network (CRAN) use
install.packages("mappoly")From GitHub (development version)
You can install the development version from Git Hub. Within R, you need to install devtools:
install.packages("devtools")If you are using Windows, you must install the the latest recommended version of Rtools.
To install MAPpoly from Git Hub use
devtools::install_github("mmollina/mappoly", dependencies=TRUE)For further QTL analysis, we recommend our QTLpoly package. QTLpoly is an under development software to map quantitative trait loci (QTL) in full-sib families of outcrossing autopolyploid species based on a random-effect multiple QTL model Pereira et al. 2020.
Workflow
Vignettes
- To access the MAPpoly vignette from R, use
vignette("mappoly_startguide") - Building a genetic map in a tetraploid potato full-sib population using MAPpoly
- Building a genetic map in an hexaploid full-sib population using MAPpoly
- Real datasets
- Hexaploid sweetpotato VCF dataset (Beauregard x Tanzania) obtained using VCF2SM
- Tetraploid potato with dosage call in MAPpoly format
- Tetraploid potato with dosage call in CSV format
- Tetraploid potato with dosage probabilities in MAPpoly format
- Tetraploid potato in CSV format obtained using ClusterCall
- Compressed tetraploid potato with dosage probabilities obtained using fitPoly
- Simulated datasets
Related software
- Polyverse - the polyploid R universe (a Lindsay Clark's initiative)
# Enable this universe
options(repos = c(
polyploids = 'https://polyploids.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
# Install some packages
install.packages('mappoly')-
Variant Calling
-
Simulations
-
Genotype calling
- ClusterCall: Automated tetraploid genotype calling by hierarchical clustering
- fitPoly: Genotype Calling for Bi-Allelic Marker Assays
- polyRAD: Genotype Calling with Uncertainty from Sequencing Data in Polyploids and Diploids
- SuperMASSA: Graphical Bayesian inference tool for genotyping polyploids
- updog: Flexible Genotyping for Polyploids
- VCF2SM: Python script that integrates VCF files and SuperMASSA
-
Genetic mapping in polyploids
-
Haplotype reconstruction
-
QTL mapping
-
Visualization
Miscellaneous
Articles referencing MAPpoly
- Using probabilistic genotypes in linkage analysis of polyploids. (Liao et al., 2021)
- Discovery of a major QTL for root-knot nematode Meloidogyne incognita resistance in cultivated sweetpotato Ipomoea batatas. (Oloka, et al., 2021)
- Quantitative trait locus mapping for common scab resistance in a tetraploid potato full-sib population. (Pereira et al., 2021)
- The recombination landscape and multiple QTL mapping in a Solanum tuberosum cv.'Atlantic'-derived F1 population. (Pereira et al., 2021)
- High-Resolution Linkage Map and QTL Analyses of Fruit Firmness in Autotetraploid Blueberry (Cappai et al., 2020)
- When a phenotype is not the genotype: Implications of phenotype misclassification and pedigree errors in genomics-assisted breeding of sweetpotato Ipomoea batatas (L.) Lam.(Gemenet et al., 2020)
- Quantitative trait loci and differential gene expression analyses reveal the genetic basis for negatively associated beta-carotene and starch content in hexaploid sweetpotato [Ipomoea batatas (L.) Lam.] (Gemenet et al., 2020)
- Multiple QTL Mapping in Autopolyploids: A Random-Effect Model Approach with Application in a Hexaploid Sweetpotato Full-Sib Population. (Pereira et al., 2020)
- Unraveling the Hexaploid Sweetpotato Inheritance Using Ultra-Dense Multilocus Mapping. (Mollinari et al., 2020).
Acknowledgment
This package has been developed as part of the Genomic Tools for Sweetpotato Improvement project (GT4SP) and SweetGAINS, both funded by Bill & Melinda Gates Foundation. Its continuous improvement is made possible by Tools for polyploids, funded by USDA NIFA Specialty Crop Research Initiative Award.
NC State University promotes equal opportunity and prohibits discrimination and harassment based upon one’s age, color, disability, gender identity, genetic information, national origin, race, religion, sex (including pregnancy), sexual orientation and veteran status.