lh3 / Minigraph
Programming Languages
Labels
Projects that are alternatives of or similar to Minigraph
Getting Started
git clone https://github.com/lh3/minigraph
cd minigraph && make
# Map sequence to sequence, similar to minimap2 without base alignment
./minigraph test/MT-human.fa test/MT-orangA.fa > out.paf
# Map sequence to graph
./minigraph test/MT.gfa test/MT-orangA.fa > out.gaf
# Incremental graph generation (-l10k necessary for this toy example)
./minigraph -xggs -l10k test/MT.gfa test/MT-chimp.fa test/MT-orangA.fa > out.gfa
# Call per-sample path in each bubble/variation
./minigraph -xasm -l10k --call test/MT.gfa test/MT-orangA.fa > orangA.call.bed
# The lossy FASTA representation (requring https://github.com/lh3/gfatools)
gfatools gfa2fa -s out.gfa > out.fa
# Extract localized structural variations
gfatools bubble out.gfa > SV.bed
Table of Contents
Introduction
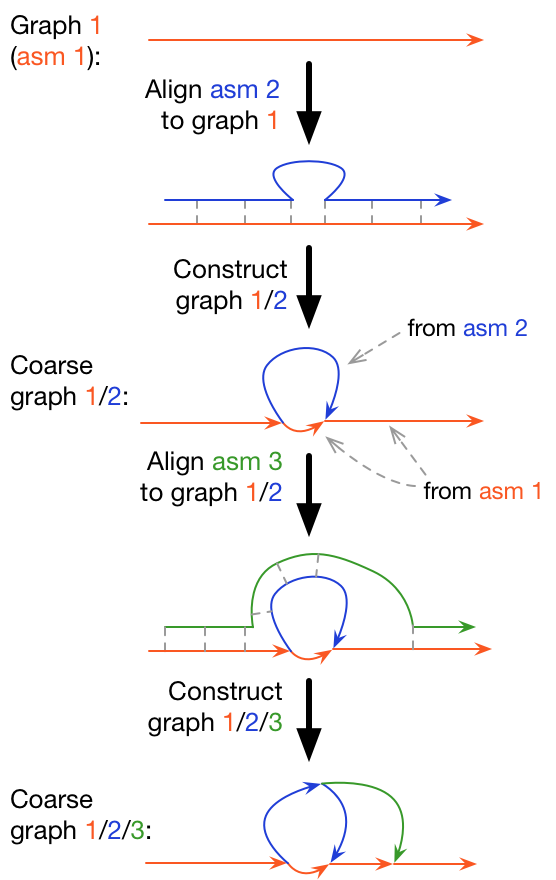
Minigraph is a sequence-to-graph mapper and graph constructor. It finds approximate locations of a query sequence in a sequence graph and incrementally augments an existing graph with long query subsequences diverged from the graph. The figure on the right briefly explains the procedure.
Minigraph borrows many ideas and code from minimap2. It is fairly efficient and can construct a graph from 40 human assemblies in half a day using 24 CPU cores. Partly due to the lack of base alignment, minigraph may produce suboptimal mappings and local graphs. Please read the Limitations section of this README for more information.
Users' Guide
Installation
To install minigraph, type make in the source code directory. The only
non-standard dependency is zlib.
Sequence-to-graph mapping
To map sequences against a graph, you should prepare the graph in the GFA format, or preferrably the rGFA format. If you don't have a graph, you can generate a graph from multiple samples (see the Graph generation section below). The typical command line for mapping is
minigraph -x lr graph.gfa query.fa > out.gaf
You may choose the right preset option -x according to input. Minigraph
output mappings in the GAF format, which is a strict superset of the
PAF format. The only visual difference between GAF and PAF is that the
6th column in GAF may encode a graph path like
>MT_human:0-4001<MT_orang:3426-3927 instead of a contig/chromosome name.
The minigraph GFA parser seamlessly parses FASTA and converts it to GFA internally, so you can also provide sequences in FASTA as the reference. In this case, minigraph will behave like minimap2 but without base-level alignment.
Graph generation
The following command-line generates a graph in rGFA:
minigraph -xggs -t16 ref.fa sample1.fa sample2.fa > out.gfa
which is equivalent to
minigraph -xggs -t16 ref.fa sample1.fa > sample1.gfa
minigraph -xggs -t16 sample1.gfa sample2.fa > out.gfa
File ref.fa is typically the reference genome (e.g. GRCh38 for human).
It can also be replaced by a graph in rGFA. Minigraph assumes sample1.fa to
be the whole-genome assembly of an individual. This is an important assumption:
minigraph only considers 1-to-1 orthogonal regions between the graph and the
individual FASTA. If you use raw reads or put multiple individual genomes in
one file, minigraph will filter out most alignments as they cover the input
graph multiple times.
The output rGFA can be converted to a FASTA file with gfatools:
gfatools gfa2fa -s graph.gfa > out.stable.fa
The output out.stable.fa will always include the initial reference ref.fa
and may additionally add new segments diverged from the initial reference.
Calling structural variations
A minigraph graph is composed of chains of bubbles with the reference as the backbone. Each bubble represents a structural variation. It can be multi-allelic if there are multiple paths through the bubble. You can extract these bubbles with
gfatools bubble graph.gfa > var.bed
The output is a BED-like file. The first three columns give the position of a bubble/variation and the rest of columns are:
- (4) # GFA segments in the bubble including the source and the sink of the bubble
- (5) # all possible paths through the bubble (not all paths present in input samples)
- (6) 1 if the bubble involves an inversion; 0 otherwise
- (7) length of the shortest path (i.e. allele) through the bubble
- (8) length of the longest path/allele through the bubble
- (9-11) please ignore
- (12) list of segments in the bubble; first for the source and last for the sink
- (13) sequence of the shortest path (
*if zero length) - (14) sequence of the longest path (NB: it may not be present in the input samples)
Given an assembly, you can find the path/allele of this assembly in each bubble with
minigraph -xasm --call graph.gfa sample-asm.fa > sample.bed
On each line in the BED-like output, the last colon separated field gives the
alignment path through the bubble, the path length in the graph, the mapping
strand of sample contig, the contig name, the approximate contig start and
contig end. The number of lines in the file is the same as the number of lines
in the output of gfatools bubble. You can use the paste Unix command to
piece multiple samples together.
Prebuilt graphs
Prebuilt human graphs in the rGFA format can be found at
ftp://ftp.dfci.harvard.edu/pub/hli/minigraph.
Algorithm overview
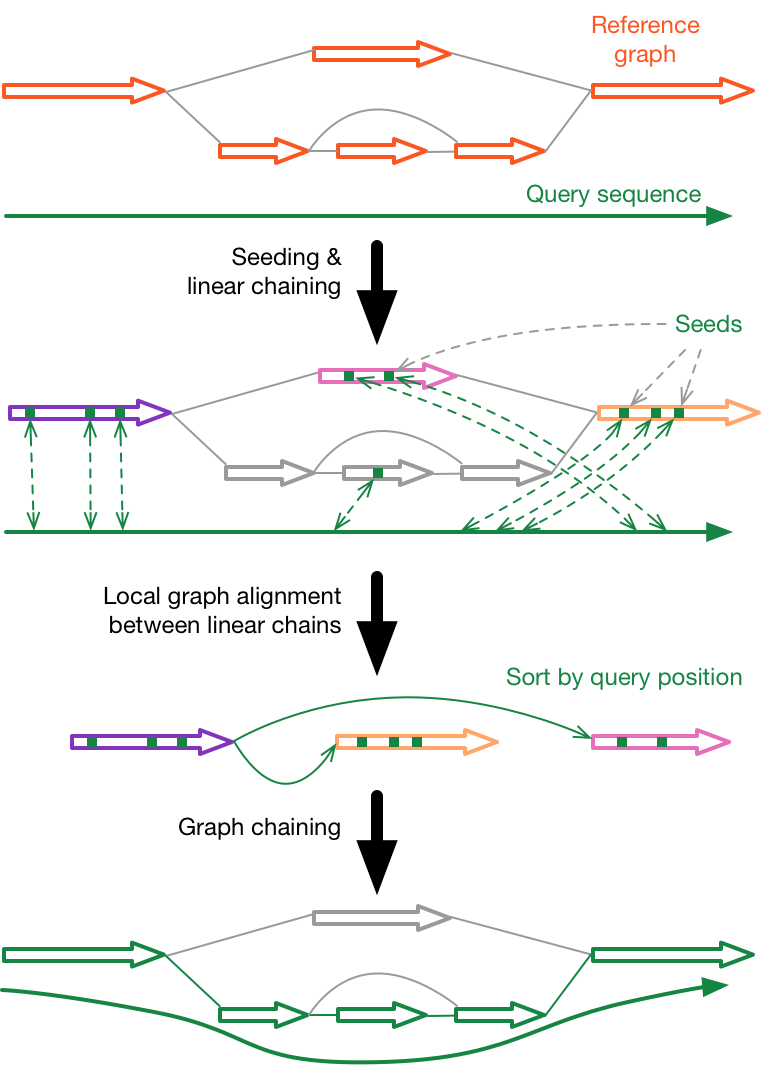
In the following, minigraph command line options have a dash ahead and are highlighted in bold. The description may help to tune minigraph parameters.
-
Read all reference bases, extract (-k,-w)-minimizers and index them in a hash table.
-
Read -K [=500M] query bases in the mapping mode, or read all query bases in the graph construction mode. For each query sequence, do step 3 through 5:
-
Find colinear minimizer chains using the minimap2 algorithm, assuming segments in the graph are disconnected. These are called linear chains.
-
Perform another round of chaining, taking each linear chain as an anchor. For a pair of linear chains, minigraph finds up to 15 shortest paths between them and chooses the path of length closest to the distance on the query sequence. Minigraph checks the base sequences, but doesn't perform thorough graph alignment. Chains found at this step are called graph chains.
-
Identify primary chains and estimate mapping quality with a method similar to the one used in minimap2.
-
In the graph construction mode, collect all mappings longer than -d [=10k] and keep their query and graph segment intervals in two lists, respectively.
-
For each mapping longer than -l [=50k], finds poorly aligned regions. A region is filtered if it overlaps two or more intervals collected at step 6.
-
Insert the remaining poorly aligned regions into the input graph. This constructs a new graph.
Limitations
-
Minigraph mainly captures length variations between samples. A complex subgraph is often suboptimal due to the lack of base alignment and the order dependency of input samples. It may not represent the evolution history or the functional relevance at the locus. Please do not overinterpret complex subgraphs. If you are interested in a particular subgraph, it is recommended to extract the input contig subsequences involved in the subgraph with the
--calloption and manually curated the results. -
Minigraph needs to find strong colinear chains first. For a graph consisting of many short segments (e.g. one generated from rare SNPs in large populations), minigraph will fail to map query sequences.
