iandanforth / Pymuscle
Programming Languages
Projects that are alternatives of or similar to Pymuscle
PyMuscle
PyMuscle provides a motor unit based model of skeletal muscle. It simulates the relationship between excitatory input and motor-unit output as well as fatigue over time.
It is compatible with OpenAI Gym environments and is intended to be useful for researchers in the machine learning community.
PyMuscle can be used to enhance the realism of motor control for simulated agents. To get you started we provide a toy example project which uses PyMuscle in a simulation of arm curl and extension.
Out of the box we provide a motor neuron pool model and a muscle fiber model based on "A motor unit-based model of muscle fatigue" (Potvin and Fuglevand, 2017). If you use this library as part of your research please cite that paper.
We hope to extend this model and support alternative models in the future.
More about PyMuscle
Motor control in biological creatures is complex. PyMuscle allows you to capture some of that complexity while remaining performant. It provides greater detail than sending torque values to simulated motors-as-joints but less detail (and computational cost) than a full biochemical model.
PyMuscle is not tied to a particular physics library and can be used with a variety of muscle body simulations. PyMuscle focuses on the relationship between control signals (excitatory inputs to motor neurons) and per-motor-unit output.
Motor unit output is dimensionless but can be interpreted as force. It can also be used as a proxy for the contractile state of muscle bodies in the physics sim of your choice.
Background
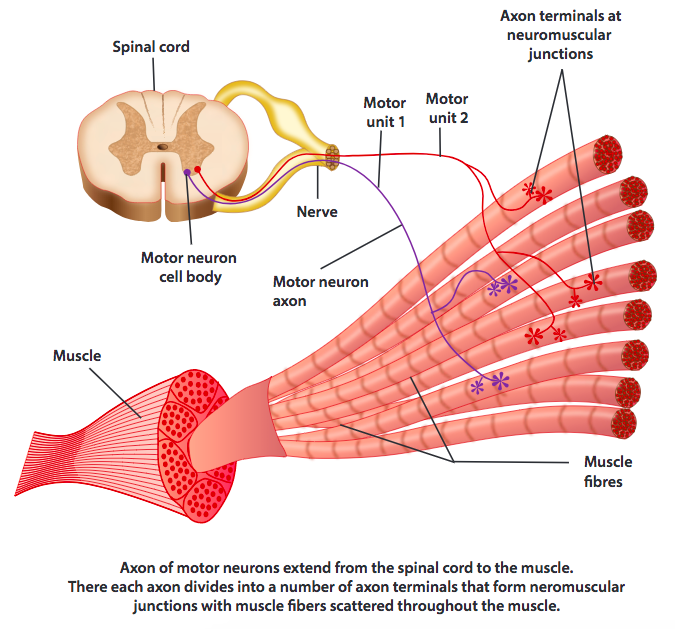
Motor Units
A motor unit is the combination of a motor neuron and the muscle fibers to which the neuron makes connections. Skeletal muscles are made up of many muscle fibers. For a given motor unit a single motor neuron will have an axon that branches and innervates a subset of the fibers in a muscle. Muscle fibers usually belong to only one motor unit.
Muscles may have anywhere from a few dozen to thousands of motor units. The human arm, for example, has 30 some muscles and is innervated by approximately 35,000 axons from motor neurons.
The brain controls muscles by sending signals to motor units and receiving signals from mechanoreceptors embedded in muscles and the skin. In animals all the motor units an animal will ever have are present from birth and learning to produce smooth coordinated motion through control of those units is a significant part of the developmental process.
Control
Motor units are recruited in an orderly fashion to produce varying levels of muscle force.
The cell bodies of motor neurons for a given muscle cluster together in the spinal cord in what are known as motor neuron pools, columns, or nuclei. Generally motor neurons in a pool can be thought of as all getting the same activation inputs. This input is the combination of dozens if not hundreds of separate inputs from interneurons and upper motor neurons carrying signals from the brain and mechanoreceptors in the body.
In a voluntary contraction of a muscle, say in curling your arm, the input to the motor neuron pool for the bicep muscle will ramp up, recruiting more and more motor units, starting from the weakest motor units to stronger ones.
Over time motor neurons and muscle fibers can't produce the same level of force for the same level of activation input. This is called fatigue. The brain must compensate for the fatigue if it wants to maintain a given force or perform the same action again and again in the same way.
Installation
Requirements
Python 3.6+
Install
pip install pymuscle
Getting Started
Not a Machine Learning researcher? Please see Getting Started for Physiologists
Minimal example
The Muscle class provides the primary API for the library. A Muscle can be heavily customized but here we use mainly default values. A PotvinMuscle instantiated with 120 motor units has the distribution of strengths, recruitment thresholds, and fatigue properties as used in the experiments of Potvin and Fuglevand, 2017.
from pymuscle import StandardMuscle as Muscle
from pymuscle.vis import PotvinChart
muscle = Muscle()
# Set up the simulation parameters
sim_duration = 200 # seconds
frames_per_second = 50
step_size = 1 / frames_per_second
total_steps = int(sim_duration / step_size)
# Use a constant level of excitation to more easily observe fatigue
excitation = 0.6 # Range is 0.0 to 1.0
total_outputs = []
outputs_by_unit = []
print("Starting simulation ...")
for i in range(total_steps):
# Calling step() updates the simulation and returns the total output
# produced by the muscle during this step for the given excitation level.
total_output = muscle.step(excitation, step_size)
total_outputs.append(total_output)
# You can also introspect the muscle to see the forces being produced
# by each motor unit.
output_by_unit = muscle.current_forces
outputs_by_unit.append(output_by_unit)
if (i % (frames_per_second * 10)) == 0:
print("Sim time - {} seconds ...".format(int(i / frames_per_second)))
# Visualize the behavior of the motor units over time
print("Creating chart ...")
chart = PotvinChart(
outputs_by_unit,
step_size
)
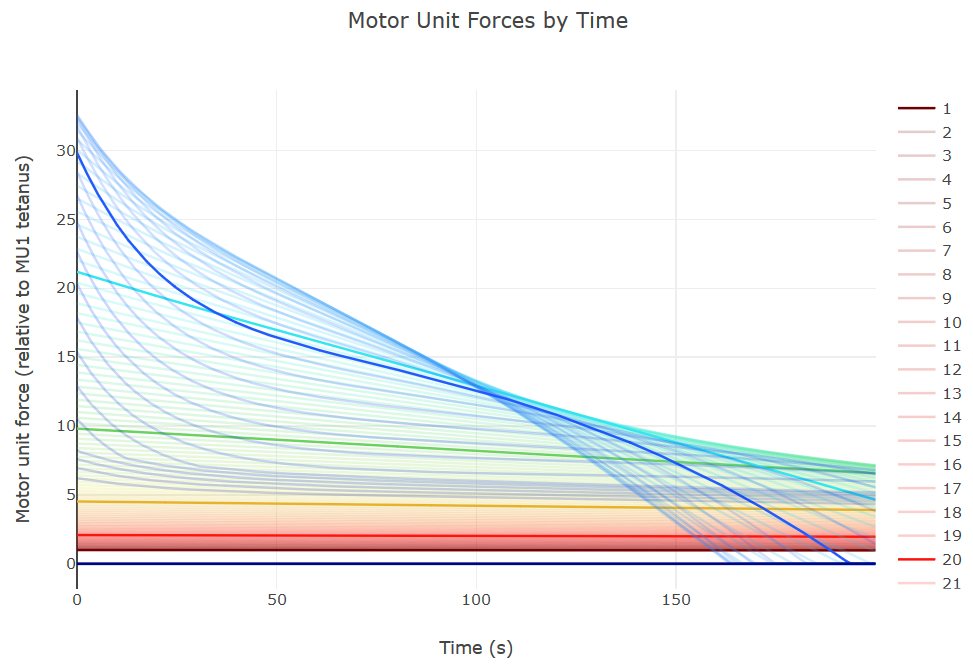
# Things to note in the chart:
# - Some motor units (purple) are never recruited at this level of excitation
# - Some motor units become completely fatigued in this short time
# - Some motor units stabilize and decrease their rate of fatigue
# - Forces from the weakest motor units are almost constant the entire time
chart.display()
This will open a browser window with the produced chart. It should look like this:
Familiar with OpenAI's Gym?
Make sure you have the following installed
pip install gym pygame pymunk
then try out the example project
Versioning Plan
PyMuscle is in an alpha state. Expect regular breaking changes.
We expect to stabilize the API for 1.0 and introduce breaking changes only during major releases.
This library tries to provide empirically plausible behavior. As new research is released or uncovered we will update the underlying model. Non-bug-fix changes that would alter the output of the library will be integrated in major releases.
If you know of results you believe should be integrated please let us know. See the Contributing section.
Contributing
We encourage you to contribute! Specifically we'd love to hear about and feature projects using PyMuscle.
For all issues please search the existing issues before submitting.
- Bug Reports
- Enhancement requests
- Suggest research that can better inform the model
Before opening a pull request please:
- See if there is an open ticket for this issue
- If the ticket is tagged 'Help Needed' comment to note that you intend to work on this issue
- If the ticket is not tagged, comment that you would like to work on the issue
- We will then discuss the priority, timing and expectations around the issue.
- If there is no open ticket, create one
- We prefer to discuss the implications of changes before you write code!
Development
If you want to help develop the PyMuscle library itself the following may help.
Clone this repository
git clone [email protected]:iandanforth/pymuscle.git
cd pymuscle
pip install -r requirements-dev.txt
python setup.py develop
pytest
Performance
PyMuscle aims to be fast. We use Numpy to get fast vector computation. If you find that some part of the library is not fast enough for your use case please open a ticket and let us know.
Limitations
Scope
PyMuscle is concerned with inputs to motor unit neurons, the outputs of those motor units, and the changes to that system over time. It does not model the dynamics of the muscle body itself or the impact of dynamic motion on this motor unit input/output relationship.
Recovery
Potvin and Fuglevand 2017 explicitly models fatigue but not recovery. We eagerly await the updated model from Potvin which will included a model of recovery.
Until then the StandardMuscle class, which builds on the Potvin and Fuglevand
base classes, implements peripheral (muscle fiber) recovery as this is a
relatively simple process but disables central (motor unit fatigue).
Proprioception
Instances of StandardMuscle implement a get_peripheral_fatigue() method
to allow users to track fatigue state of each muscle. Other than that this
library does not directly provide any feedback signals for control. The
example projects show how to integrate PyMuscle with a physics simulation to
get simulated output forces and stretch and strain values derived from the
state of the simulated muscle body. (In the example this is a damped spring
but a Hill-type, or more complex model could also be used.)