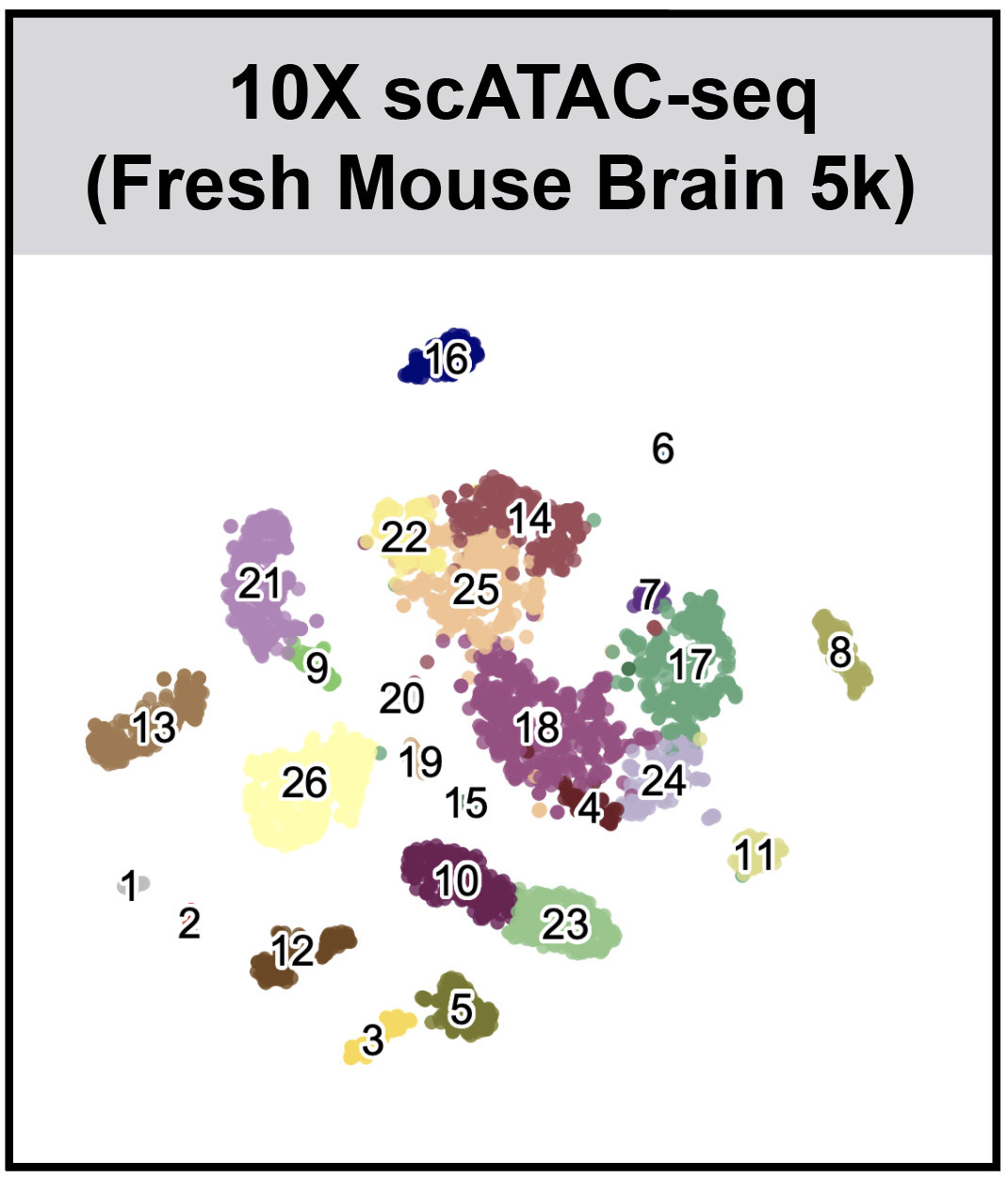
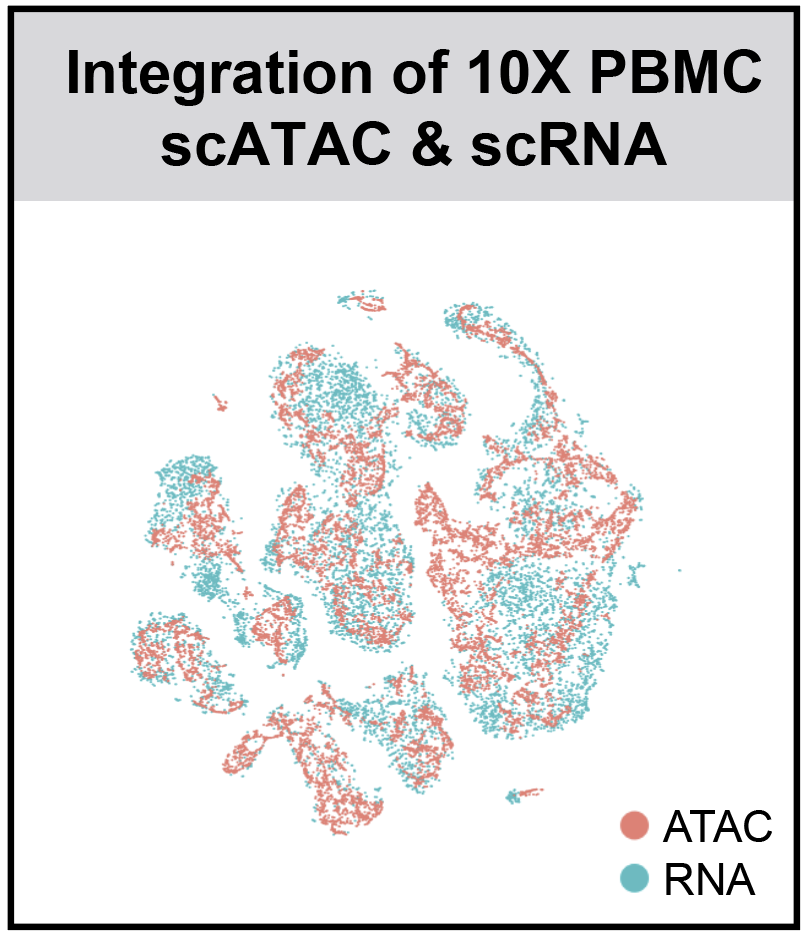
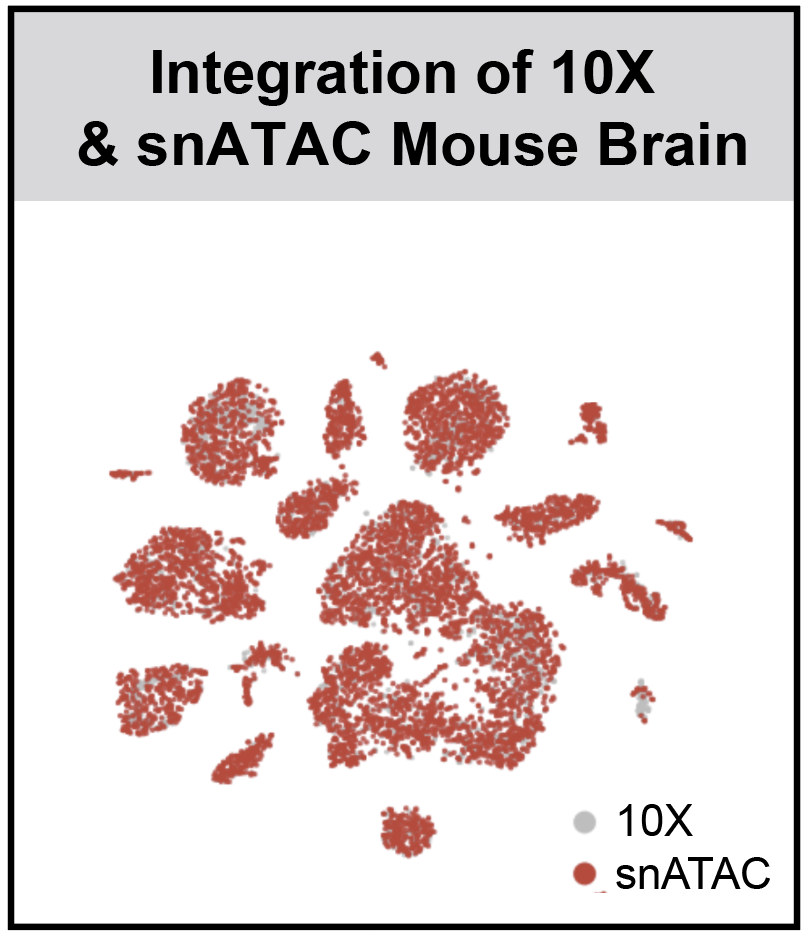
r3fang / Snapatac
Programming Languages
Projects that are alternatives of or similar to Snapatac
SnapATAC (Latest Updates: 2019-09-19)
SnapATAC (Single Nucleus Analysis Pipeline for ATAC-seq) is a fast, accurate and comprehensive method for analyzing single cell ATAC-seq datasets.
Latest News
- SnapATAC links distal elements to putative target genes
- SnapATAC integrates scRNA and scATAC
- SnapATAC employs a new method for dimensionality reduction
- SnapATAC enables clustering using leiden algorithm
- SnapATAC enables batch effect correction
- SnapATAC enables motif analysis using chromVAR
FAQs
- How to run SnapATAC on 10X dataset?
- I already ran CellRanger, can I use its output for SnapATAC?
- How can I analyze combine multiple samples together?
- How to group reads from any subset of cells?
- What is a snap file?
- How to create a snap file from fastq file?
Requirements
- Linux/Unix
- Python (>= 2.7 & < 3.0) (SnapTools) (highly recommanded for 2.7);
- R (>= 3.4.0 & < 3.6.0) (SnapATAC) (3.6 does not work for rhdf5 package);
Pre-print
Rongxin Fang, Sebastian Preissl, Xiaomeng Hou, Jacinta Lucero, Xinxin Wang, Amir Motamedi, Andrew K. Shiau, Eran A. Mukamel, Yanxiao Zhang, M. Margarita Behrens, Joseph Ecker, Bing Ren. Fast and Accurate Clustering of Single Cell Epigenomes Reveals Cis-Regulatory Elements in Rare Cell Types. bioRxiv 615179; doi: https://doi.org/10.1101/615179
Installation
SnapATAC has two components: Snaptools and SnapATAC.
- SnapTools - a python module for pre-processing and working with snap file.
- SnapATAC - a R package for the clustering, annotation, motif discovery and downstream analysis.
Install snaptools from PyPI. See how to install snaptools on FAQs. NOTE: Please use python 2.7 if possible.
$ pip install snaptools
Install SnapATAC R pakcage (development version).
$ R
> library(devtools)
> install_github("r3fang/SnapATAC")