Nellaker-group / Towardsdeepphenotyping
Labels
Projects that are alternatives of or similar to Towardsdeepphenotyping
Towards Deep Placental Histology Phenotyping
@article{ferlaino2018towards,
title={Towards Deep Cellular Phenotyping in Placental Histology},
author={Ferlaino, Michael and Glastonbury, Craig A and Motta-Mejia, Carolina and Vatish, Manu and Granne, Ingrid and Kennedy, Stephen and Lindgren, Cecilia M and Nell{\aa}ker, Christoffer},
journal={arXiv preprint arXiv:1804.03270},
year={2018}
}
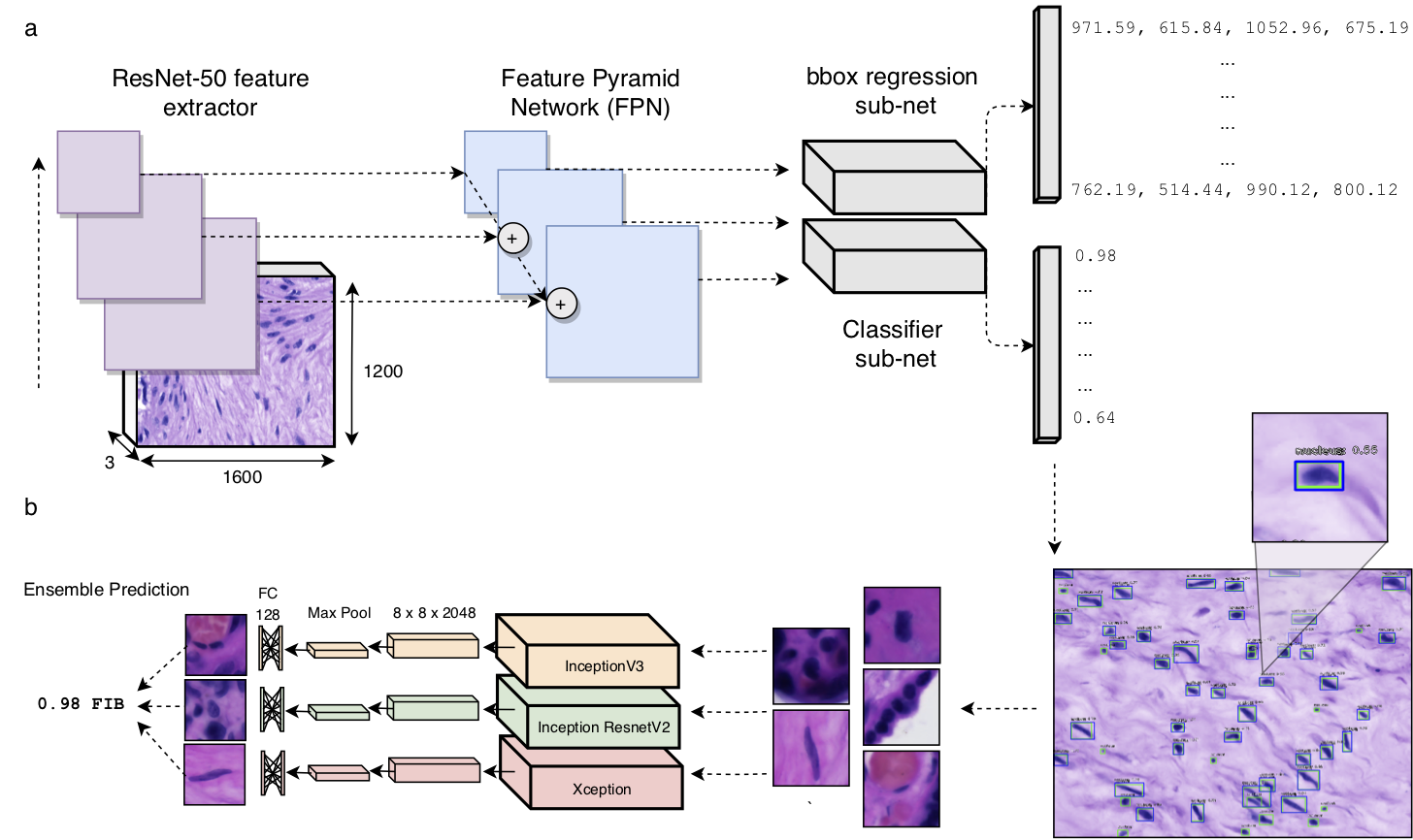
Nuclei Detector
All annotations are available in this repository along with training, validation and test images.
To train RetinaNet with COCO-pretrained weights, run:
keras_retinanet/bin/train.py --epochs 100 --weights resnet50_coco_best_v1.2.2.h5 --steps 71 --batch-size 1 csv train_nuclei_annotations.csv class_mapping.txt --val-annotations valid_nuclei_annotations.csv
To evaluate our model (resnet50_csv_37.h5) on the test images, run:
keras_retinanet/bin/evaluate.py --max-detections 500 --score-threshold 0.50 --save-path detections/ csv test_nuclei_annotations.csv class_mapping.txt ./snapshots/resnet50_csv_37.h5
A notebook is provided (Evaluate_RetinaNet.ipynb) to further validate this model on an additional set of test images (14K tiles).
Cell Classification
We trained an ensemble system to stratify placental cells into 5 distinct populations. We used 3 base classifiers (InceptionV3, InceptionResNetV2, and Xception), which were fined tuned on our data set of histological images.
Add data used to fine tune cell classifiers can be found in the folder "Datasets/CellClassifierData/".
Furthermore, the scripts used to train, validate, and test all base classifers are collected in "FineTuningScripts/". The training for each of the base learner is identical (barring batch sizes). For instance, to fine tune InceptionV3, run:
python FineTuneInceptionV3.py --save=True
The command trains InceptionV3, saves training logs (by default the flag "save" is False), and stores the best model (as assessed by validation accuracy).
For more details, please refer to our arXiv paper:
Towards Deep Cellular Phenotyping in Placental Histology
Pipeline Deployment on Whole Images
In the folder "Notebooks", Pipeline.ipynb displays an example of analysis and visualisations that can be generated by deploying our pipeline for predictions across whole images.
Model Weights and Benchmark Data
Additional test images (19GB for about 14K images), that can be used as benchmark data, are provided here.
All models, fine tuned on our histological data sets, are freely available for download using the following links:
| Model | Weights |
|---|---|
| RetinaNet | 500MB |
| FCNN | 20MB |
| InceptionV3 | 199MB |
| InceptionResNetV2 | 440MB |
| Xception | 209MB |