ykwon0407 / Uq_bnn
Labels
Projects that are alternatives of or similar to Uq bnn
Uncertainty quantification using Bayesian neural networks in classification: Application to biomedical imaging segmentation
This repository provides the Keras implementation of the paper "Uncertainty quantification using Bayesian neural networks in classification: Application to biomedical image segmentation" at the Computational Statistics and Data Analysis. This paper extends the paper accepted at the MIDL 2018. In case you want to cite this work, please cite the extended version.
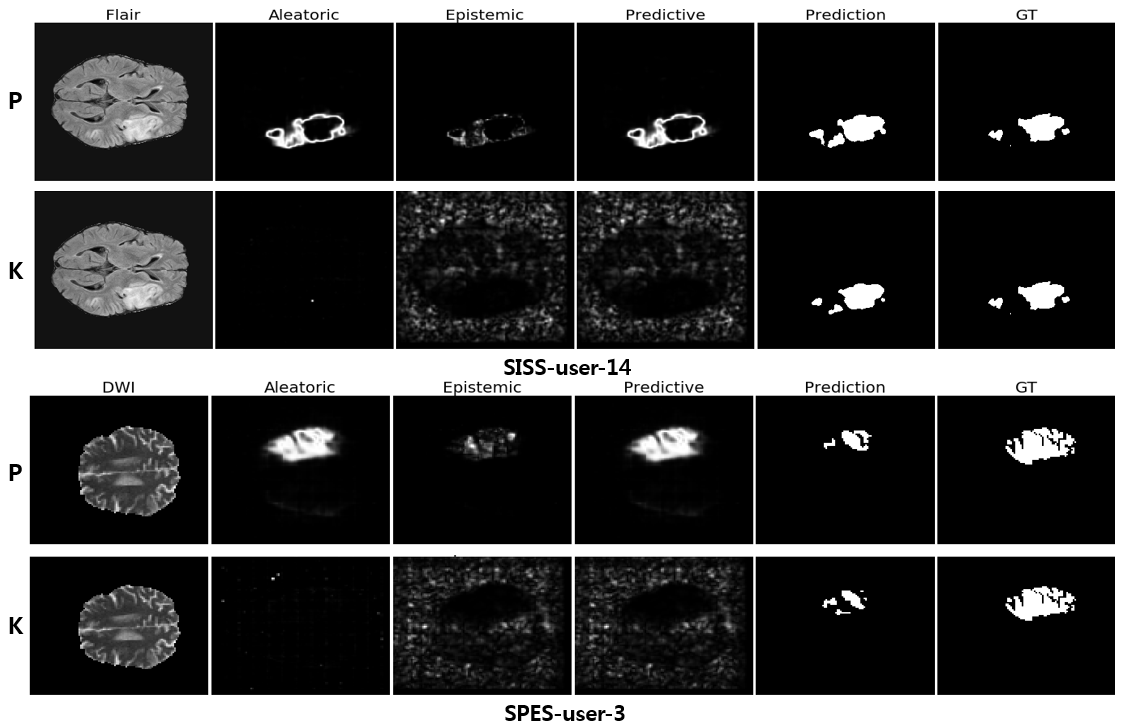
In this repo, we demonstrate the proposed method using the two biomedical imaging segmentation datasets: the ISLES and the DRIVE datasets. For more detailed information, please see the ISLES and DRIVE.
I also strongly recommend to see the good implementation using the DRIVE dataset by Walter de Back. [notebook].
Example
Once you have a trained Bayesian neural network, the proposed uncertainty quantification method is simple !!! In a binary segmentaion, a numpy array p_hat with dimension (number of estimates, dimension of features), then the epistemic and aleatoric uncertainties can be obtained by the following code.
epistemic = np.mean(p_hat**2, axis=0) - np.mean(p_hat, axis=0)**2
aleatoric = np.mean(p_hat*(1-p_hat), axis=0)
A directory tree
.
├── ischemic
│ ├── input
│ │ └── (train/test datasets)
│ └── src
│ ├── configs (empty)
│ ├── data.py
│ ├── models.py
│ ├── settings.py
│ ├── train.py
│ ├── utils.py
│ └── weights (empty)
├── README.md
└── retina
├── fig
├── input
│ └── (train/test datasets)
├── model.py
├── UQ_DRIVE_stochastic_sample_2000.ipynb
├── UQ_DRIVE_stochastic_sample_200.ipynb
├── utils.py
└── weights (empty)
References
- ISLES website
- DRIVE website
- Oskar Maier et al. ISLES 2015 - A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI, Medical Image Analysis, Available online 21 July 2016, ISSN 1361-8415, http://dx.doi.org/10.1016/j.media.2016.07.009.
- J.J. Staal, M.D. Abramoff, M. Niemeijer, M.A. Viergever, B. van Ginneken, "Ridge based vessel segmentation in color images of the retina", IEEE Transactions on Medical Imaging, 2004, vol. 23, pp. 501-509.
- Kendall, Alex, and Yarin Gal. "What uncertainties do we need in bayesian deep learning for computer vision?." Advances in neural information processing systems. 2017.
Author
Yongchan Kwon, Ph.D. student, Department of Statistics, Seoul National University