j-duan / Vs Net
Programming Languages
Projects that are alternatives of or similar to Vs Net
VS-Net: Variable splitting network for accelerated parallel MRI reconstruction
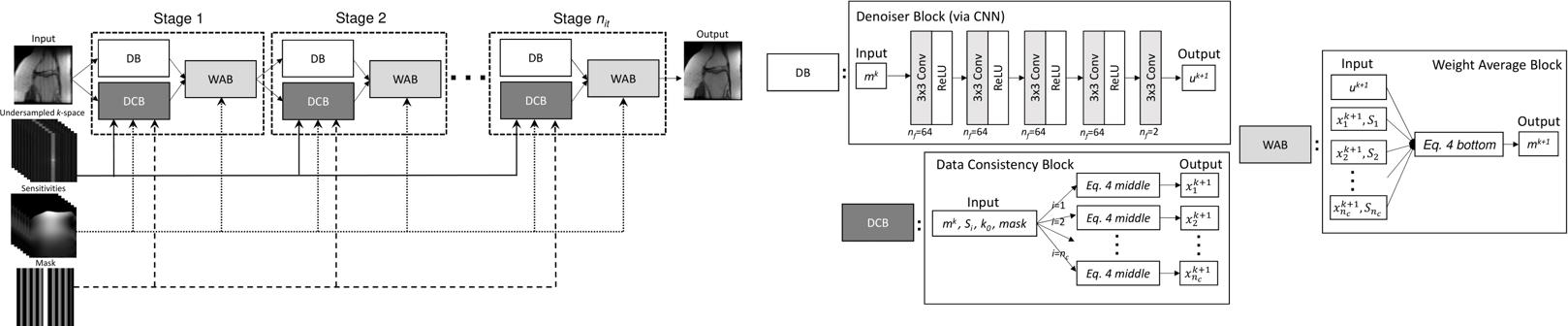
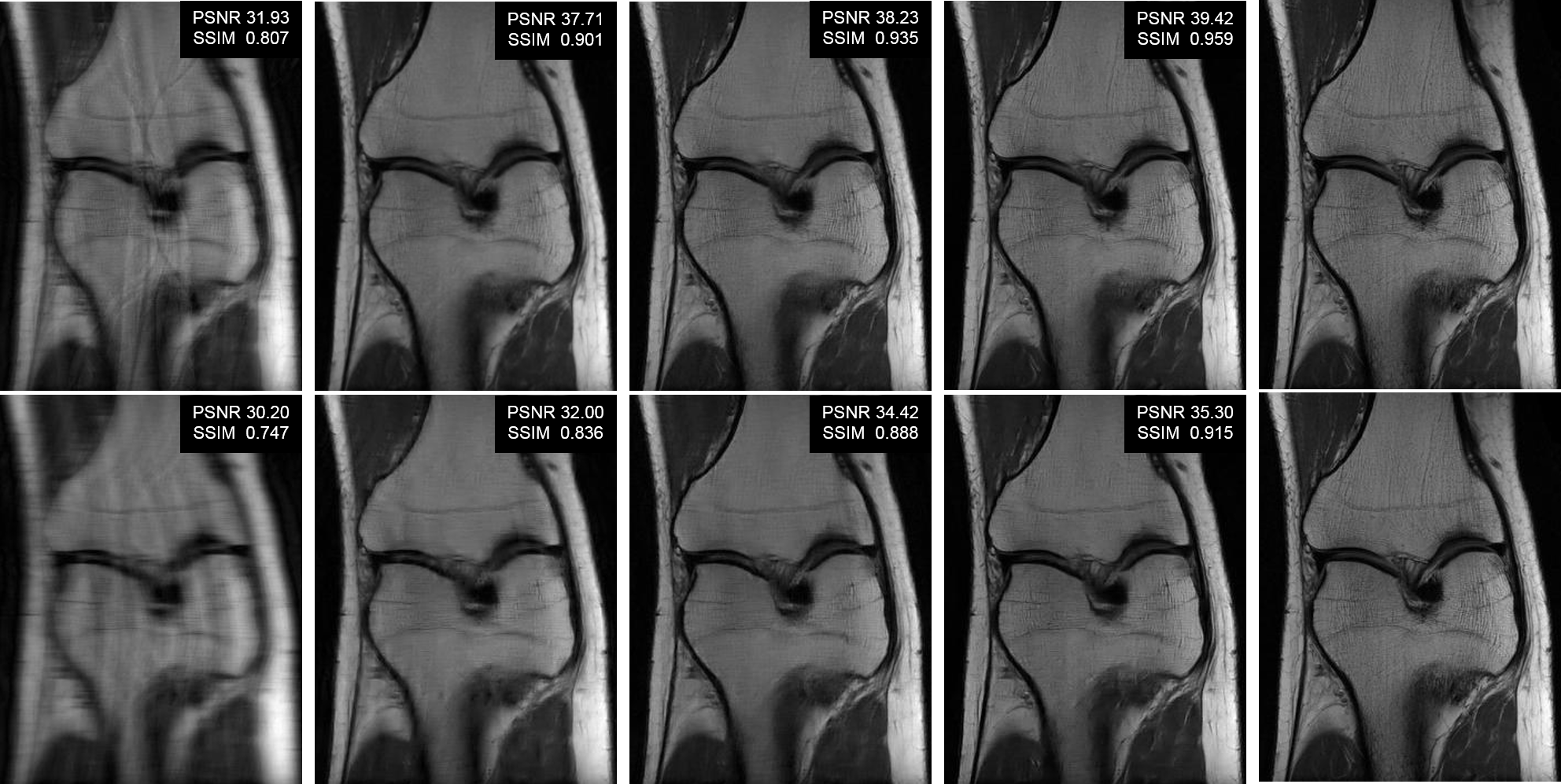
The code in this repository implements VS-Net (Fig.1), a model-driven neural network for accelerated parallel MRI reconstruction (see our presentation slides and poster). Specifically, we formulate the generalized parallel compressed sensing reconstruction as an energy minimization problem, for which a variable splitting optimization method is derived. Based on this formulation we propose a novel, end-to-end trainable deep neural network architecture by unrolling the resulting iterative process of such variable splitting scheme. We evaluated VS-Net on complex valued multi-coil knee images for 4-fold and 6-fold acceleration factors showed improved performance (Fig.2).
 |
|---|
| Fig.1: VS-Net overall architecture (left) and each block in VS-net (right). DB, DCB and WAB stand for Denoiser Block, Data Consistency Block and Weighted Average Block, respectively. |
 |
|---|
| Fig.2: Visual comparison using Cartesian undersampling with AF 4 (top) and 6 (bottom). From left to right: zero-filling, l1-SPIRiT, Variational Network, VS-Net and ground truth. Click here for more visual comparison. |
Overview
The files in this repository are organized into 5 directories and 1 root directory:
- root : contains base functions for training, validation, inference and visualization:
- network architecture, as shown in Fig.1 - architecture.py
- data loader to read complex-valued raw MRI data and sensitivity maps - data_loader.py
- inference to deploy a trained model on unseen raw data - inference.py
- save png images for visualization after inference - save_png.py
- train and validate the VS-Net - vs_net.py
- common : contains dependant functions used in training or deploying VS-Net and is written by fastMRI with some of our modificatins
- data : contains dependant functions used in training or deploying VS-Net and is written by fastMRI
-
log : produces
csvfiles where the quantitative metrics (PSNR, SSIM and NMSE) over each iteration are saved - model : saves trained models. There are 4 pre-trained models that can be used directly to see VS-Net performance.
-
results : save final results. After inference is run, this folder will produce 3 mat files, i.e.
vs-200.mat,zero_filling.matandreference.mat. On top of the three mat files, running save_png.py will produce png files.
Run VS-Net
To start the training process with vs_net.py, please follow the following four steps:
1. Download the knee data
Download the data we used for our experiments at GLOBUS.
2. Install python libraries
pip install visdom torch==1.2.1 matplotlib h5py scipy scikit-image
3. Start visdom
python -m visdom.server
4. Run vs_net.py
you need to change the path in this python script to where you save the knee data downloaded above. For visualization during training, open your browser and enter http://localhost:8097
Citation
If you find this software useful for your project or research. Please give some credits to authors who developed it by citing the following paper. We really appreciate that. If you encounter any problem during installation, please feel free to contact me via [email protected]
[1] Duan J, Schlemper J, Qin C, Ouyang C, Bai W, Biffi C, Bello G, Statton B, O'Regan DP, Rueckert D. VS-Net: Variable splitting network for accelerated parallel MRI reconstruction. arXiv preprint arXiv:1907.10033. MICCAI (2019).
