tkuanlun350 / 3dunet Tensorflow Brats18
Programming Languages
Projects that are alternatives of or similar to 3dunet Tensorflow Brats18
3DUnet-Tensorflow
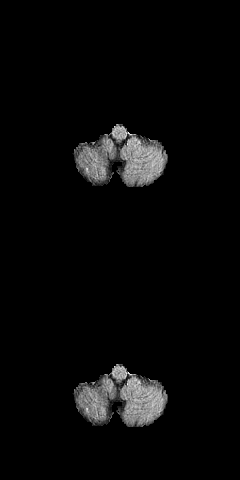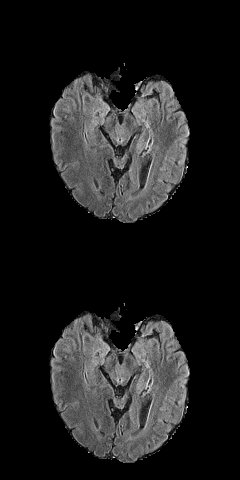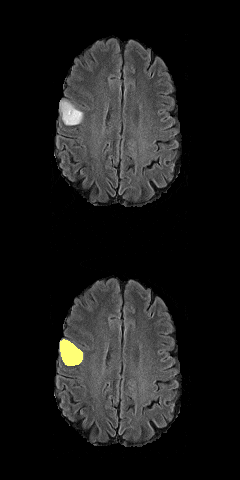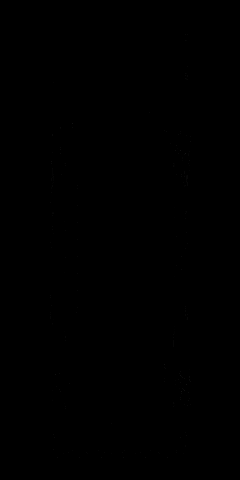
3D Unet biomedical segmentation model powered by tensorpack with fast io speed.
Borrow a lot of codes from https://github.com/taigw/brats17/. I improved the pipeline and using tensorpack's dataflow for faster io speed. Currently it takes around 7 minutes for 500 iterations with patch size [5 X 20 X 144 X 144]. You can achieve reasonable results within 40 epochs (more gpu will also reduce your training time.)
I want to verify the effectiveness (consistent improvement despite of slight implementation differences and different deep-learning framework) of some architecture proposed these years. Such as dice_loss, generalised dice_loss, residual connection, instance norm, deep supervision ...etc. Those design are popular and used in many papers in BRATS competition.
- BRATS2017 1rd Solution, Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation
- BRATS2017 2rd Solution, Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks
- BRATS2017 3rd Solution
Dependencies
- Python 3; TensorFlow >= 1.4
- [email protected] (https://github.com/tensorpack/tensorpack) (pip install -U git+https://github.com/ppwwyyxx/[email protected])
- BRATS2017 or BRATS2018 data. It needs to have the following directory structure (you can also add your own validation data and test data, just to modify the path in config):
- (Optional) If you want to use Bias Correction you have to install nipype and ANTs (see preprocess.py)
DIR/
training/
HGG/
LGG/
val/
BRATS*.nii.gz
Data
If you don't have Brats data, you can visit ellisdg/3DUnetCNN where he provided sample data from TCGA.
You can modify data_loader.py to apply for different 3D datasets. The data sampling strategy is defined in data_sampler.py BatchData class.
Usage
Change config in config.py:
- Change
BASEDIRto/path/to/DIRas described above.
Train:
python3 train.py --logdir=./train_log/unet3d --gpu 0
Eval:
python3 train.py --load=./train_log/unet3d/model-30000 --gpu 0 --evaluate
Predict:
python3 train.py --load=./train_log/unet3d/model-30000 --gpu 0 --predict
If you want to use 5 fold cross validation :
- Run generate_5fold.py to save 5fold.pkl
- Set config CROSS_VALIDATION to True
- Set config CROSS_VALIDATION_PATH to {/path/to/5fold.pkl}
- Set config FOLD to {0~4}
Results
The detailed parameters and training settings. The results are derived from Brats2018 online evaluation on Validation Set.
Single Model
Setting 1:
Unet3d, num_filters=32 (all), depth=3, sampling=one_positive
- PatchSize = [5, 20, 144, 144] per gpu, num_gpus = 2, epochs = 40
- Lr = 0.01, num_step=500, epoch time = 6:35(min), total_training_time ~ 5 hours
Setting 2:
Unet3d, num_filters=32 (all), depth=3, sampling=one_positive
- PatchSize = [2, 128, 128, 128] pre gpu, num_gpus = 2, epochs = 40
- Lr = 0.01, num_step=500, epoch time = 20:35(min), total_training_time ~ 8 hours
Setting 3
Unet3d, num_filters=16~256, sampling=one_positive, depth=5, residual
- PatchSize = [2, 128, 128, 128], num_gpus = 1, epochs = 20
- Lr = 0.001, num_step=500, epoch time = 20(min), total_training_time ~ 8 hours
Setting 4:
Unet3d, num_filters=16~256, depth=5, residual InstanceNorm, sampling=random
- PatchSize = [2, 128, 128, 128], num_gpus = 1, epochs = 20
- Lr = 0.001, num_step=500, epoch time = 20(min), total_training_time ~ 8 hours
Setting 5:
Unet3d, num_filters=16~256, depth=5, residual, InstanceNorm, sampling=one_positive
- PatchSize = [2, 128, 128, 128], num_gpus = 1, epochs = 20
- Lr = 0.001, num_step=500, epoch time = 20(min), total_training_time ~ 8 hours
Setting 6:
Unet3d, num_filters=16~256, depth=5, residual, deep-supervision, InstanceNorm, sampling=one_positive
- PatchSize = [2, 128, 128, 128], num_gpus = 1, epochs = 20
- Lr = 0.001, epoch time = 19(min), total_training_time ~ 8 hours
Setting 7:
Unet3d, num_filters=16~256, depth=5, residual, deep-supervision, BatchNorm, sampling=one_positive
- PatchSize = [2, 128, 128, 128], num_gpus = 1, epochs = 20
- Lr = 0.001, epoch time = 20(min), total_training_time ~ 8 hours
Setting 8:
Unet3d, num_filters=16~256, depth=5, residual, deep-supervision, InstanceNorm, sampling=random
- PatchSize = [2, 128, 128, 128], num_gpus = 2, epochs = 20
- Lr = 0.001, epoch time = 22(min), total_training_time ~ 8 hours
| Setting | Dice_ET | Dice_WT | Dice_TC |
|---|---|---|---|
| 1 | 0.74 | 0.85 | 0.75 |
| 2 | 0.74 | 0.83 | 0.77 |
| 2* | 0.77 | 0.84 | 0.77 |
| 3 | 0.74 | 0.87 | 0.78 |
| 4 | 0.75 | 0.87 | 0.790 |
| 5 | 0.72 | 0.87 | 0.796 |
| 6 | 0.73 | 0.88 | 0.80 |
| 6* | 0.75 | 0.88 | 0.80 |
| 7 | 0.73 | 0.87 | 0.78 |
| 8* | 0.77 | 0.87 | 0.81 |
Ensemble Results
Multi-View:
Introduced by Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks. Trained with axial, sagittal and coronal and then average the prediction prob.
Currently only support manually set path for each model (see train.py after line 147.)
Test-Time augmentation:
Testing with image augmentation to improve model robustness.
- Flip: Predicting on original image and horizontal flipped image and average the prediction prob.
| Setting | Dice_ET | Dice_WT | Dice_TC |
|---|---|---|---|
| 8+Flip | 0.73 | 0.88 | 0.81 |
| 8*+Flip | 0.77 | 0.88 | 0.82 |
| Multi-View* | 0.78 | 0.89 | 0.81 |
| Multi-View*+Flip | 0.78 | 0.89 | 0.82 |
p.s. * means advanced post-processing
Preprocessing
Zero Mean Unit Variance (default)
Normalize each modality with zero mean and unit variance within brain region
Bias Correction
| Setting | Dice_ET | Dice_WT | Dice_TC |
|---|---|---|---|
| N4+8*+Flip | 0.76 | 0.87 | 0.80 |
| Multi-View*+N4+Flip | 0.76 | 0.89 | 0.80 |
Using preprocess.py to convert Brats data into corrected image. Will take one days to process 200+ files. (multi-threading could help)
Notes
Results for brats2018 will be updated and more experiments will be included. [2018/8/3]