wikke / Tianchi Medical Lungtumordetect
天池医疗AI大赛[第一季]:肺部结节智能诊断 UNet/VGG/Inception/ResNet/DenseNet
Stars: ✭ 314
Projects that are alternatives of or similar to Tianchi Medical Lungtumordetect
Keras Idiomatic Programmer
Books, Presentations, Workshops, Notebook Labs, and Model Zoo for Software Engineers and Data Scientists wanting to learn the TF.Keras Machine Learning framework
Stars: ✭ 720 (+129.3%)
Mutual labels: jupyter-notebook, resnet, densenet, inception, vgg
Caffe Model
Caffe models (including classification, detection and segmentation) and deploy files for famouse networks
Stars: ✭ 1,258 (+300.64%)
Mutual labels: classification, segmentation, resnet, inception
Tensornets
High level network definitions with pre-trained weights in TensorFlow
Stars: ✭ 982 (+212.74%)
Mutual labels: resnet, densenet, inception, vgg
Medicalzoopytorch
A pytorch-based deep learning framework for multi-modal 2D/3D medical image segmentation
Stars: ✭ 546 (+73.89%)
Mutual labels: segmentation, resnet, unet, densenet
Paddlex
PaddlePaddle End-to-End Development Toolkit(『飞桨』深度学习全流程开发工具)
Stars: ✭ 3,399 (+982.48%)
Mutual labels: classification, segmentation, resnet, unet
Segmentation models
Segmentation models with pretrained backbones. Keras and TensorFlow Keras.
Stars: ✭ 3,575 (+1038.54%)
Mutual labels: segmentation, resnet, unet, densenet
Pytorch Speech Commands
Speech commands recognition with PyTorch
Stars: ✭ 128 (-59.24%)
Mutual labels: classification, resnet, densenet
Unet
unet for image segmentation
Stars: ✭ 3,751 (+1094.59%)
Mutual labels: jupyter-notebook, segmentation, unet
Pytorch classification
利用pytorch实现图像分类的一个完整的代码,训练,预测,TTA,模型融合,模型部署,cnn提取特征,svm或者随机森林等进行分类,模型蒸馏,一个完整的代码
Stars: ✭ 395 (+25.8%)
Mutual labels: jupyter-notebook, resnet, densenet
Food Recipe Cnn
food image to recipe with deep convolutional neural networks.
Stars: ✭ 448 (+42.68%)
Mutual labels: jupyter-notebook, classification, vgg
Pytorch Image Classification
Tutorials on how to implement a few key architectures for image classification using PyTorch and TorchVision.
Stars: ✭ 272 (-13.38%)
Mutual labels: jupyter-notebook, resnet, vgg
Segmentation
Tensorflow implementation : U-net and FCN with global convolution
Stars: ✭ 101 (-67.83%)
Mutual labels: classification, segmentation, unet
Pytorch Classification
Classification with PyTorch.
Stars: ✭ 1,268 (+303.82%)
Mutual labels: classification, resnet, densenet
LibtorchTutorials
This is a code repository for pytorch c++ (or libtorch) tutorial.
Stars: ✭ 463 (+47.45%)
Mutual labels: vgg, resnet, unet
Dlcv for beginners
《深度学习与计算机视觉》配套代码
Stars: ✭ 1,244 (+296.18%)
Mutual labels: classification, segmentation, unet
python cv AI ML
用python做计算机视觉,人工智能,机器学习,深度学习等
Stars: ✭ 73 (-76.75%)
Mutual labels: vgg, densenet, resnet
Multiclass Semantic Segmentation Camvid
Tensorflow 2 implementation of complete pipeline for multiclass image semantic segmentation using UNet, SegNet and FCN32 architectures on Cambridge-driving Labeled Video Database (CamVid) dataset.
Stars: ✭ 67 (-78.66%)
Mutual labels: jupyter-notebook, segmentation, unet
阿里云天池医疗大赛·肺结节检测
Features
- 3D Segmentation & Classification with Keras
- Fine preprocessing with scikit-image
- Fine visualization for clarification
- Modified UNet for segmentation
- Modified VGG/Inception/ResNet/DenseNet for classification ensemble
- Fine hyperparameter tunning with both models and training process.
Code Hierarchy
- config.py # good practice to centralize hyper parameters
- preprocess.py # Step 1, preprocess, store numpy/meta 'cache' at ./preprocess/
- train_segmentation.py # Step 2, segmentation with UNet Model
- model_UNet.py # UNet model definition
- train_classificaion.py # Step 3, classificaiton with VGG/Inception/ResNet/DenseNet
- model_VGG.py # VGG model definition
- model_Inception.py # Inception model definition
- model_ResNet.py # ResNet model definition
- model_DenseNet.py # DenseNet model definition
- generators.py # generator for segmentation & classificaiton models
- visual_utils.py # 3D visual tools
- dataset/ # dataset, changed in config.py
- preprocess/ # 'cache' preprocessed numpy/meta data, changed in config.py
- train_ipynbs # training process notebooks
Preprocess
- use
SimpleITKto read CT files, process, and store into cache with numpy arrays - process with
scikit-imagelib, try lots of parameters for best cutting- binarized
- clear-board
- label
- regions
- closing
- dilation
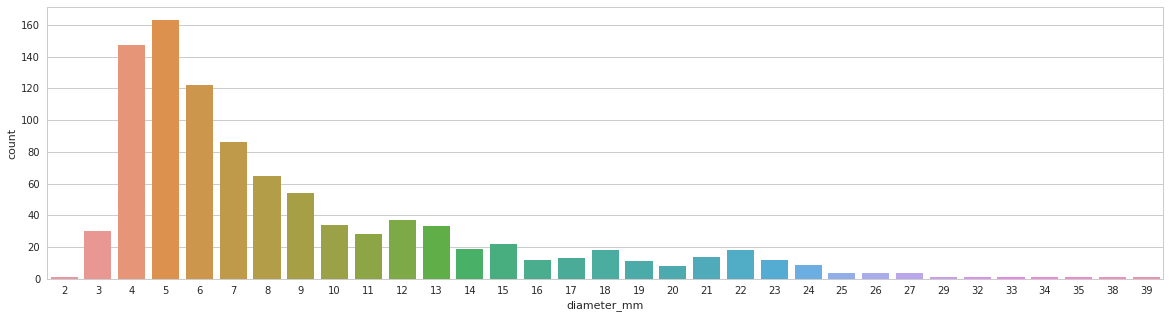
- collect all meta information(seriesuid, shape, file_path, origin, spacing, coordinates, cover_ratio, etc.) and store in ONE cache file for fast training init.
- see preprocessing in
/train_ipynbs/preprocess.ipynbfile
Distribution of the lung part takes on a whole CT.
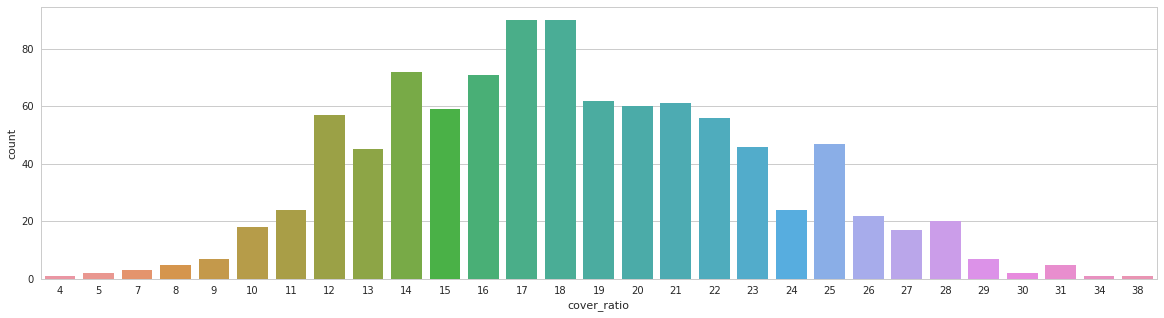
Tumor size distribution
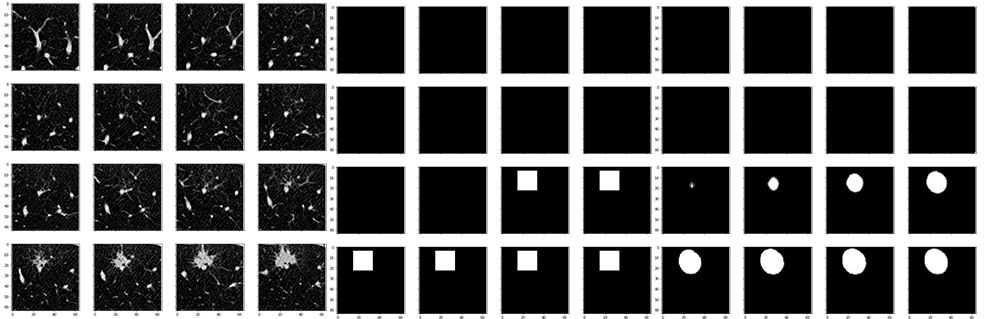
Segmentation
- A simplified and full UNet both tested.
-
dice_coef_lossas loss function. - Periodically evaluate model with lots of metrics, which helps a lot to understand the model.
- 30% of negative sample, which has no tumor, for generalization.
- Due to memory limitation, 16 batch size used.
Classification
VGG
- A simplified and full VGG model both tested. Use simplified VGG as baseline.
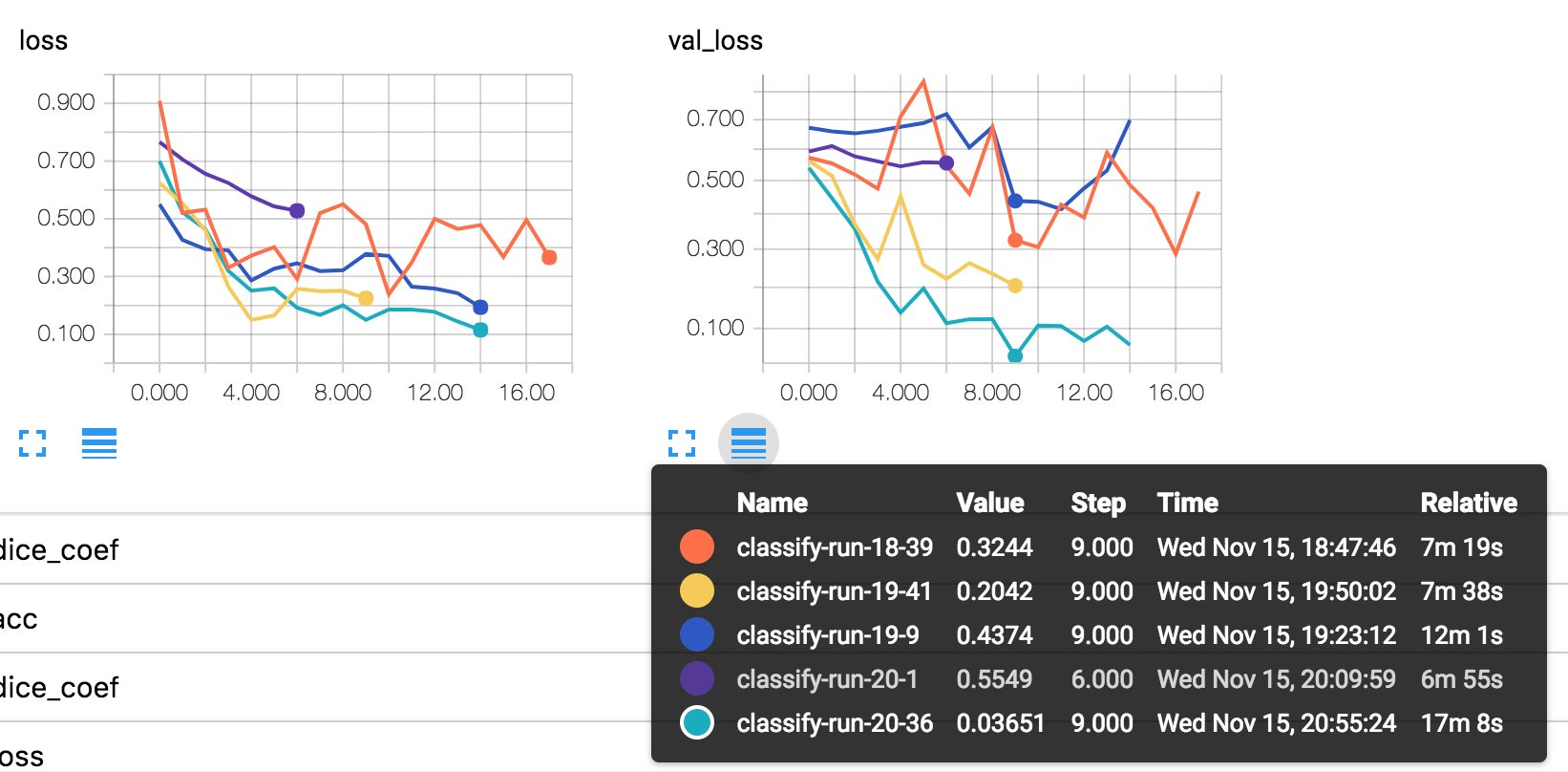
Pictures tells that: hyperparameter tunning really matters.
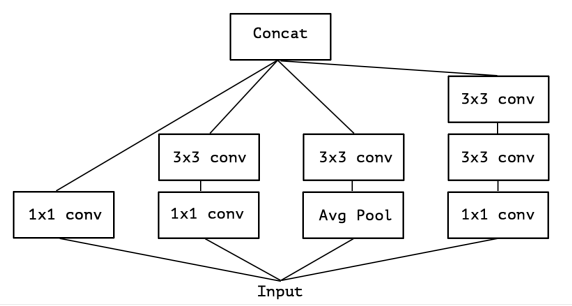
Inception
- A simplified Inception-module based network, with each block has 4-5 different type of conv.
- 1*1*1 depth-size seperable conv
- 1*1*1 depth-size seperable conv, then 3*3*3 conv_bn_relu
- 1*1*1 depth-size seperable conv, then 2 3*3*3 conv_bn_relu
- AveragePooling3D, then 1*1*1 depth-size seperable conv
- (optional in config) 1*1*1 depth-size seperable conv, and (5, 1, 1), (1, 5, 1), (1, 1, 5) spatial separable convolution
- Concatenate above.
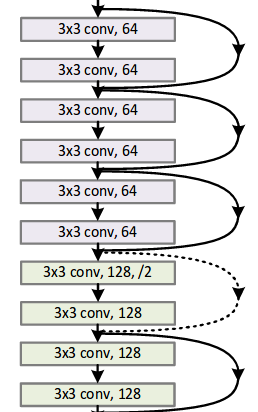
ResNet
- use
bottleneckblock instead ofbasic_blockfor implementation. - A
bottleneckresidual block consists of:- (1, 1, 1) conv_bn_relu
- (3, 3, 3) conv_bn_relu
- (1, 1, 1) conv_bn_relu
- (optional in config) kernel_size=(3, 3, 3), strides=(2, 2, 2) conv_bn_relu for compression.
- Add(not Concatenate) with input
- Leave
RESNET_BLOCKSas config to tune
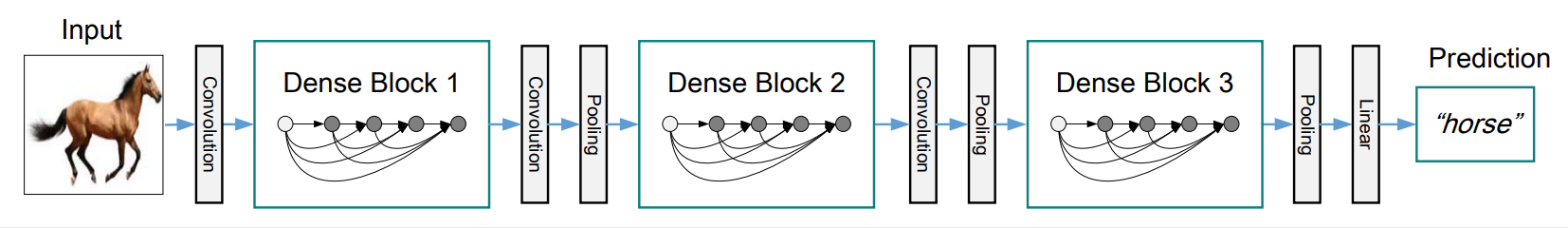
DenseNet
-
DenseNetdraws tons of experience from origin paper. https://arxiv.org/abs/1608.06993- 3 dense_block with 5 bn_relu_conv layers according to paper.
- transition_block after every dense_block, expcet the last one.
- Optional config for DenseNet-BC(paper called it): 1*1*1 depth-size seperable conv, and transition_block compression.
Fine Tunning & Experience Got
- Learning rate:
3e-5works well for UNet,1e-4works well for classification models. - Due to memory limitation, 16 batch size used.
- Data Augumentation: shift, rotate, etc.
- Visualization cannot be more important!!!
- coord(x, y, z) accord to (width, height, depth), naughty bugs.
- Put all config in one file save tons of time. Make everything clean and tidy
- Disk read is bottle neck. Read from SSD.
- Different runs has different running log dirs, for better TensorBoard visualization. Make it like
/train_logs/<model-name>-run-<hour>-<minute>. - Lots of debug options in config file.
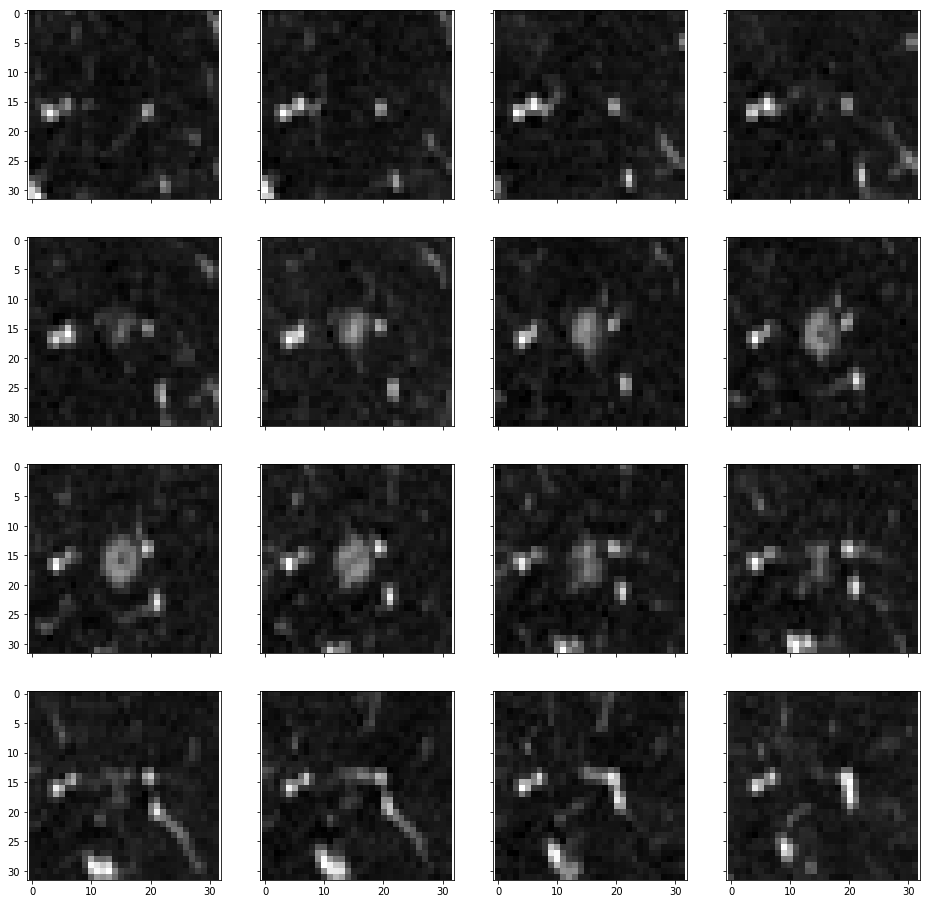
- 4 times probability strengthened for tumors < 10mm, 3 for tumor > 10mm and < 30mm, keep for > 30mm. Give more focus on small tumors, like below.
Note that the project description data, including the texts, logos, images, and/or trademarks,
for each open source project belongs to its rightful owner.
If you wish to add or remove any projects, please contact us at [email protected].
