vuptran / Cardiac Segmentation
Programming Languages
Projects that are alternatives of or similar to Cardiac Segmentation
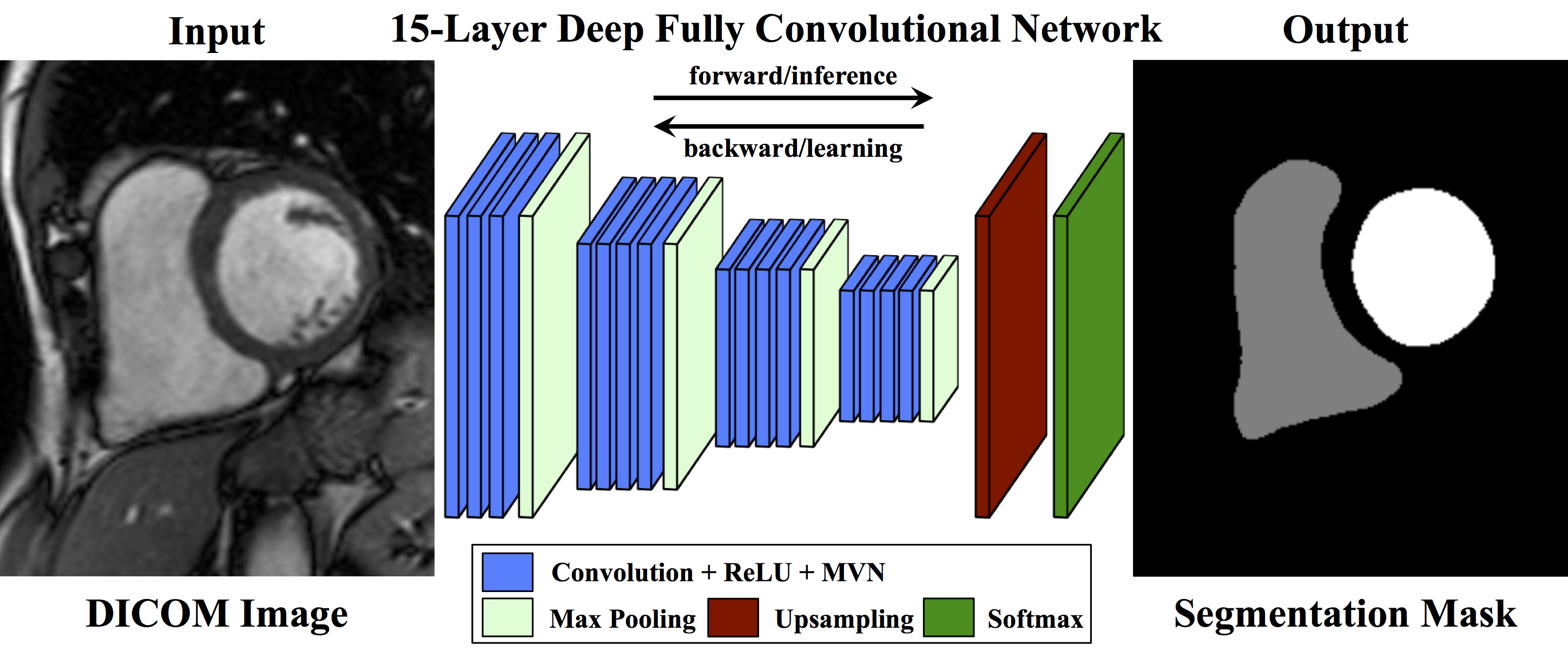
A Fully Convolutional Neural Network for Cardiac Segmentation
A Keras re-implementation of the original Caffe FCN model in the arXiv paper A Fully Convolutional Neural Network for Cardiac Segmentation in Short-Axis MRI .
Care was taken to reproduce the results reported in the original paper, particularly Tables 2-4. However, there are key differences between this Keras implementation and the original Caffe implementation:
- Caffe has the Net::Reshape method that "allows networks to change their input sizes in-place." This method is very useful for defining a fully convolutional network that can process inputs with variable shapes. Some Keras layers (such as
Cropping2DorFlatten) require shape information, and thus do not work with such variable input shapes. For this Keras FCN model, we standardize all inputs to a fixed shape, and then transform them back to their original shapes during post-processing. Caffe FCN models can process any variable input shape. - The Caffe implementation uses cross entropy loss as the training signal, whereas this Keras implementation uses the Dice coefficient as the training loss.
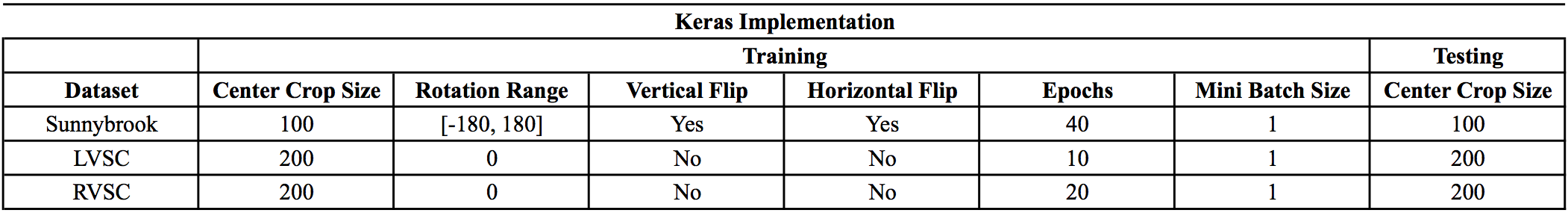
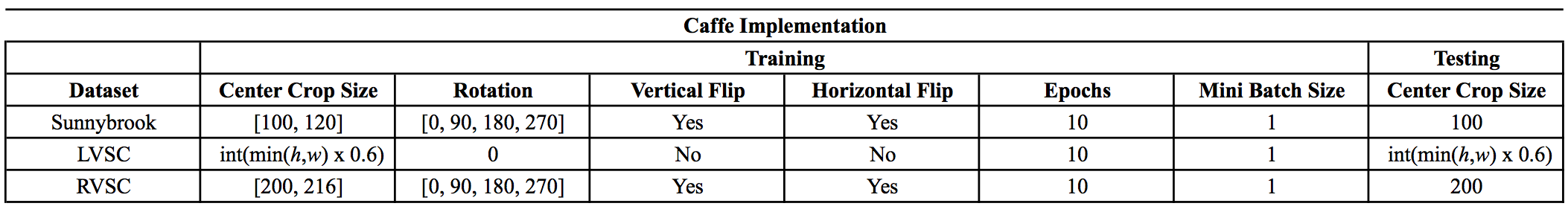
- The Caffe implementation uses a slightly different strategy for data augmentation than this Keras implementation. The following tables summarize the data augmentation and training protocol for each dataset:
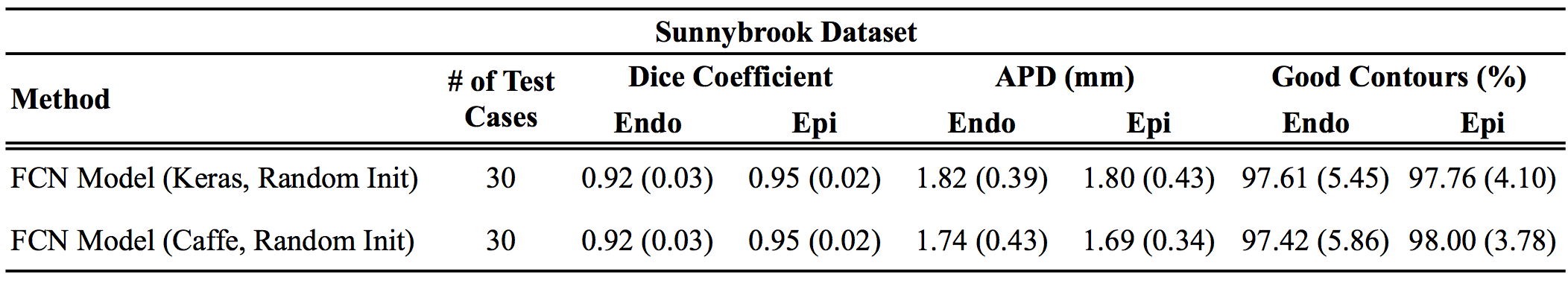
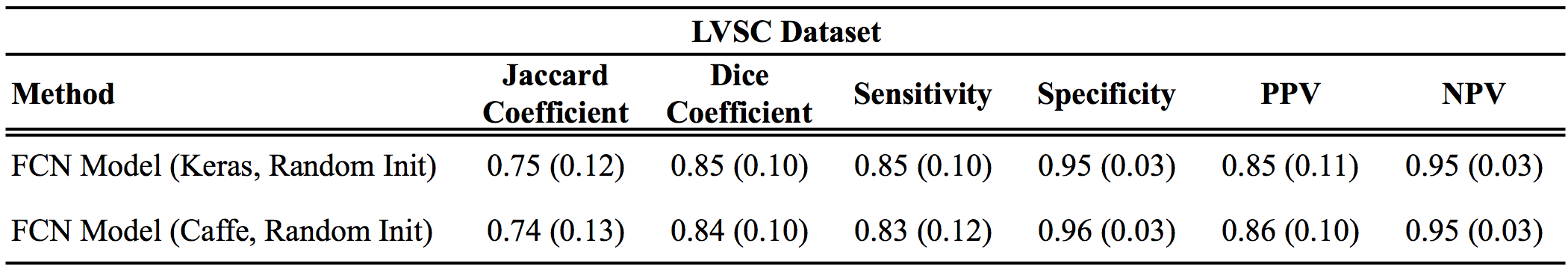
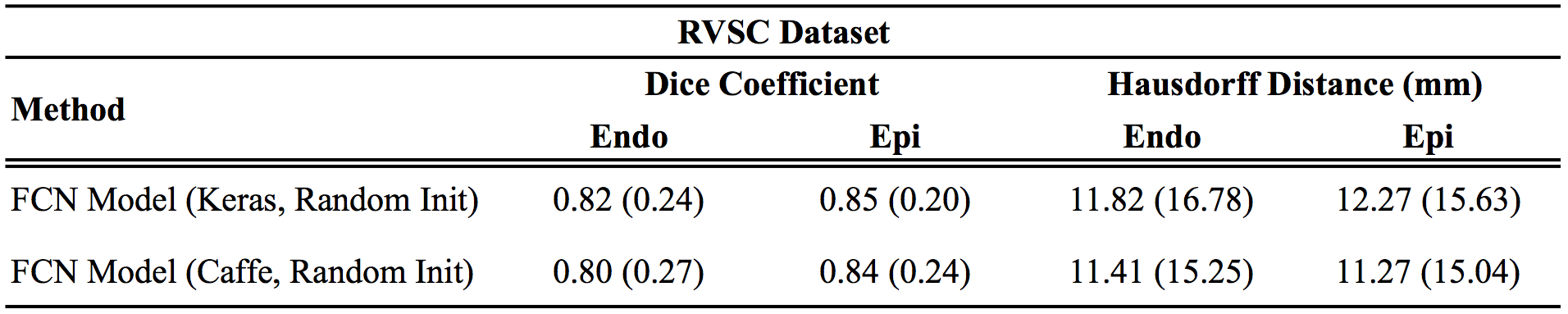
Results
Below are the Keras results as compared to the original Caffe results reported in the paper:
For all metrics, larger values are better, except for distance metrics (APD and Hausdorff), where smaller values indicate better results.
Requirements
The code is tested on Ubuntu 14.04 with the following components:
Software
- Python 2.7
- Keras 2.0.2 using TensorFlow GPU 1.0.1 backend
- CUDA 8.0 with CuDNN 5.1
- OpenCV 3.1
- h5py 2.7
- NumPy 1.11
- PyDicom 0.9.9
- Scikit-Image 0.13
Datasets
Usage
For training and evaluation, execute the following in the same directory where the datasets reside:
# Train the FCN model on the Sunnybrook dataset
$ python train_sunnybrook.py <i/o> <gpu_id>
# Train the FCN model on the LVSC dataset
$ python train_lvsc.py <i/o/myo> <gpu_id>
# Train the FCN model on the RVSC dataset
$ python train_rvsc.py <i/o> <gpu_id>
The flag <i/o/myo> indicates inner endocardium, outer epicardium, and myocardium contours, respectively, and <gpu_id> denotes the GPU device ID.
To create submission files for the test sets, execute the following:
# Create submission files for the Sunnybrook dataset
$ bash create_submission_sunnybrook.sh <gpu_id>
# Create submission files for the LVSC dataset
$ bash create_submission_lvsc.sh <gpu_id>
# Create submission files for the RVSC dataset
$ bash create_submission_rvsc.sh <gpu_id>
Note: The LVSC and RVSC submission files must be submitted to the respective LVSC and RVSC challenge organizers for the official results evaluation. The Sunnybrook submission files can be evaluated using the MATLAB code provided as part of the data download.