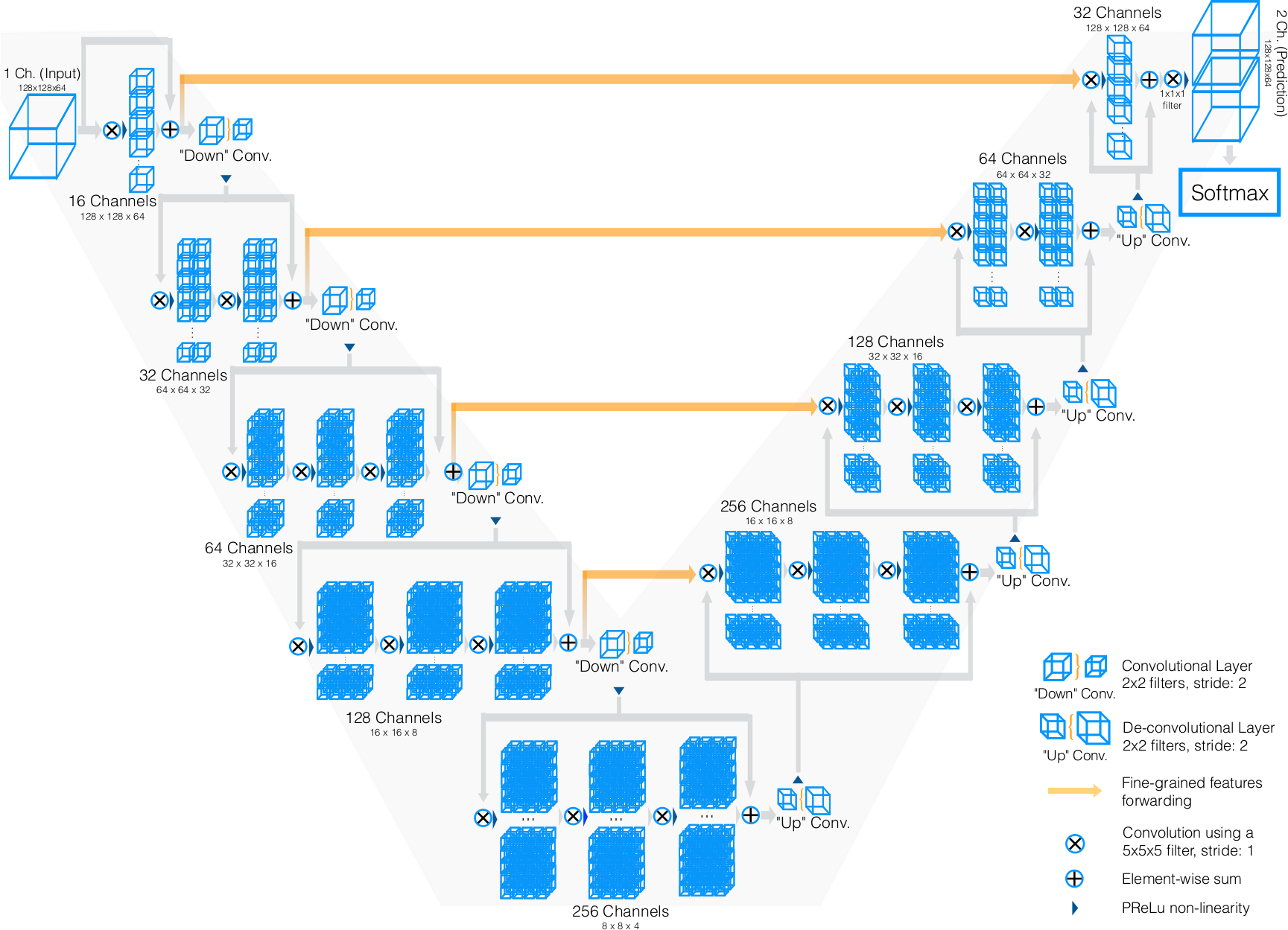
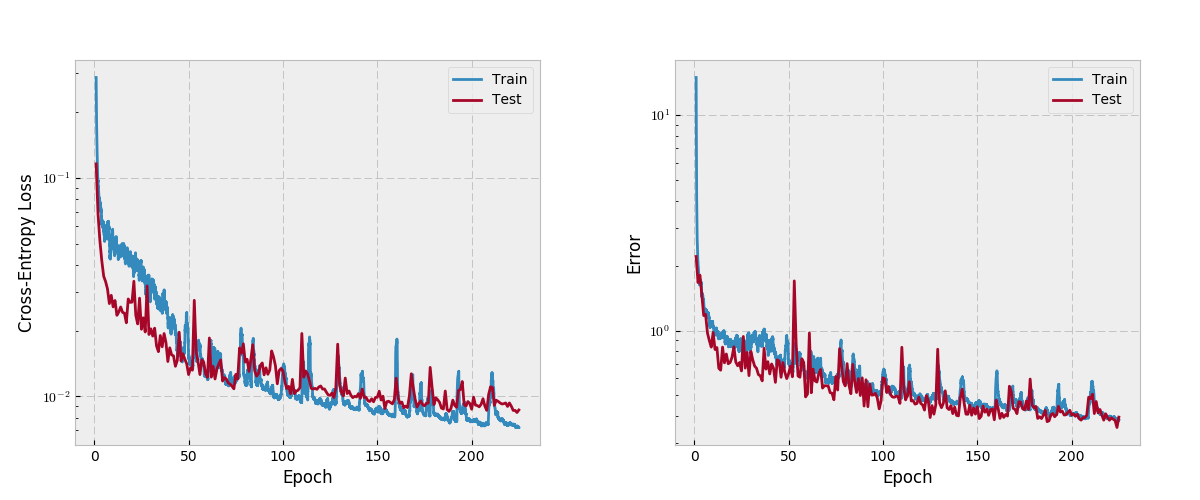
3dunet abdomen cascade Stars: ✭ 91 (-82.02%)
Mutual labels: convolutional-neural-networks, semantic-segmentation, fully-convolutional-networks
semantic segmentationSemantically segment the road in the given image.
Stars: ✭ 91 (-82.02%)
Mutual labels: semantic-segmentation, fully-convolutional-networks
atomaiDeep and Machine Learning for Microscopy
Stars: ✭ 77 (-84.78%)
Mutual labels: semantic-segmentation, fully-convolutional-networks
EspnetESPNet: Efficient Spatial Pyramid of Dilated Convolutions for Semantic Segmentation
Stars: ✭ 473 (-6.52%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
YnetY-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images
Stars: ✭ 100 (-80.24%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
Self Driving Golf CartBe Driven 🚘
Stars: ✭ 147 (-70.95%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
BonnetBonnet: An Open-Source Training and Deployment Framework for Semantic Segmentation in Robotics.
Stars: ✭ 274 (-45.85%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
Chainer PspnetPSPNet in Chainer
Stars: ✭ 76 (-84.98%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
Geospatial Machine LearningA curated list of resources focused on Machine Learning in Geospatial Data Science.
Stars: ✭ 289 (-42.89%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
Pytorch SemsegSemantic Segmentation Architectures Implemented in PyTorch
Stars: ✭ 3,180 (+528.46%)
Mutual labels: semantic-segmentation, fully-convolutional-networks
Cascaded FcnSource code for the MICCAI 2016 Paper "Automatic Liver and Lesion Segmentation in CT Using Cascaded Fully Convolutional NeuralNetworks and 3D Conditional Random Fields"
Stars: ✭ 296 (-41.5%)
Mutual labels: semantic-segmentation, fully-convolutional-networks
Cardiac SegmentationConvolutional Neural Networks for Cardiac Segmentation
Stars: ✭ 94 (-81.42%)
Mutual labels: convolutional-neural-networks, fully-convolutional-networks
Espnetv2A light-weight, power efficient, and general purpose convolutional neural network
Stars: ✭ 377 (-25.49%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
Fashion-Clothing-ParsingFCN, U-Net models implementation in TensorFlow for fashion clothing parsing
Stars: ✭ 29 (-94.27%)
Mutual labels: semantic-segmentation, fully-convolutional-networks
Elektronn3A PyTorch-based library for working with 3D and 2D convolutional neural networks, with focus on semantic segmentation of volumetric biomedical image data
Stars: ✭ 78 (-84.58%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
FCNN-exampleThis is a fully convolutional neural net exercise to detect houses from aerial images.
Stars: ✭ 28 (-94.47%)
Mutual labels: semantic-segmentation, fully-convolutional-networks
MinkowskiengineMinkowski Engine is an auto-diff neural network library for high-dimensional sparse tensors
Stars: ✭ 1,110 (+119.37%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
Espnetv2 CoremlSemantic segmentation on iPhone using ESPNetv2
Stars: ✭ 66 (-86.96%)
Mutual labels: convolutional-neural-networks, semantic-segmentation
Fcn Pytorch🚘 Easiest Fully Convolutional Networks
Stars: ✭ 278 (-45.06%)
Mutual labels: semantic-segmentation, fully-convolutional-networks
Bcdu NetBCDU-Net : Medical Image Segmentation
Stars: ✭ 314 (-37.94%)
Mutual labels: convolutional-neural-networks, semantic-segmentation