fasiha / Ebisu
Programming Languages
Projects that are alternatives of or similar to Ebisu
Ebisu: intelligent quiz scheduling
Important links
Table of contents
- Ebisu: intelligent quiz scheduling
Introduction
Consider a student memorizing a set of facts.
- Which facts need reviewing?
- How does the student’s performance on a review change the fact’s future review schedule?
Ebisu is a public-domain library that answers these two questions. It is intended to be used by software developers writing quiz apps, and provides a simple API to deal with these two aspects of scheduling quizzes:
-
predictRecallgives the current recall probability for a given fact. -
updateRecalladjusts the belief about future recall probability given a quiz result.
Behind these two simple functions, Ebisu is using a simple yet powerful model of forgetting, a model that is founded on Bayesian statistics and exponential forgetting.
With this system, quiz applications can move away from “daily review piles” caused by less flexible scheduling algorithms. For instance, a student might have only five minutes to study today; an app using Ebisu can ensure that only the facts most in danger of being forgotten are reviewed.
Ebisu also enables apps to provide an infinite stream of quizzes for students who are cramming. Thus, Ebisu intelligently handles over-reviewing as well as under-reviewing.
This document is a literate source: it contains a detailed mathematical description of the underlying algorithm as well as source code for a Python implementation (requires Scipy and Numpy). Separate implementations in JavaScript (Ebisu.js), Java (ebisu-java) and Dart (ebisu_dart) exist.
The next section is a Quickstart guide to setup and usage. See this if you know you want to use Ebisu in your app.
Then in the How It Works section, I contrast Ebisu to other scheduling algorithms and describe, non-technically, why you should use it.
Then there’s a long Math section that details Ebisu’s algorithm mathematically. If you like Beta-distributed random variables, conjugate priors, and marginalization, this is for you. You’ll also find the key formulas that implement predictRecall and updateRecall here.
Nerdy details in a nutshell: Ebisu begins by positing a Beta prior on recall probability at a certain time. As time passes, the recall probability decays exponentially, and Ebisu handles that nonlinearity exactly and analytically—it requires only a few Beta function evaluations to predict the current recall probability. Next, a quiz is modeled as a binomial trial whose underlying probability prior is this non-conjugate nonlinearly-transformed Beta. Ebisu approximates the non-standard posterior with a new Beta distribution by matching its mean and variance, which are also analytically tractable, and require a few evaluations of the Beta function.
Finally, the Source Code section presents the literate source of the library, including several tests to validate the math.
Quickstart
Install pip install ebisu (both Python3 and Python2 ok 🤠).
Data model For each fact in your quiz app, you store a model representing a prior distribution. This is a 3-tuple: (alpha, beta, t) and you can create a default model for all newly learned facts with ebisu.defaultModel. (As detailed in the Choice of initial model parameters section, alpha and beta define a Beta distribution on this fact’s recall probability t time units after it’s most recent review.)
Predict a fact’s current recall probability ebisu.predictRecall(prior: tuple, tnow: float) -> float where prior is this fact’s model and tnow is the current time elapsed since this fact’s most recent review. tnow may be any unit of time, as long as it is consistent with the half life's unit of time. The value returned by predictRecall is a probability between 0 and 1.
Update a fact’s model with quiz results ebisu.updateRecall(prior: tuple, success: int, total: int, tnow: float) -> tuple where prior and tnow are as above, and where success is the number of times the student successfully exercised this memory during the current review session out of total times—this way your quiz app can review the same fact multiple times in one sitting! The returned value is this fact’s new prior model—the old one can be discarded.
IPython Notebook crash course For a conversational introduction to the API in the context of a mocked quiz app, see this IPython Notebook crash course.
Further information Module docstrings in a pinch but full details plus literate source below, under Source code.
Alternative implementations Ebisu.js is a JavaScript port for browser and Node.js. ebisu-java is for Java and JVM languages. ebisu_dart is a Dart port for browser and native targets.
How it works
There are many scheduling schemes, e.g.,
- Anki, an open-source Python flashcard app (and a closed-source mobile app),
- the SuperMemo family of algorithms (Anki’s is a derivative of SM-2),
- Memrise.com, a closed-source webapp,
- Duolingo has published a blog entry and a conference paper/code repo on their half-life regression technique,
- the Leitner and Pimsleur spacing schemes (also discussed in some length in Duolingo’s paper).
- Also worth noting is Michael Mozer’s team’s Bayesian multiscale models, e.g., Mozer, Pashler, Cepeda, Lindsey, and Vul’s 2009 NIPS paper and subsequent work.
Many of these are inspired by Hermann Ebbinghaus’ discovery of the exponential forgetting curve, published in 1885, when he was thirty-five. He memorized random consonant–vowel–consonant trigrams (‘PED’, e.g.) and found, among other things, that his recall decayed exponentially with some time-constant.
Anki and SuperMemo use carefully-tuned mechanical rules to schedule a fact’s future review immediately after its current review. The rules can get complicated—I wrote a little field guide to Anki’s, with links to the source code—since they are optimized to minimize daily review time while maximizing retention. However, because each fact has simply a date of next review, these algorithms do not gracefully accommodate over- or under-reviewing. Even when used as prescribed, they can schedule many facts for review on one day but few on others. (I must note that all three of these issues—over-reviewing (cramming), under-reviewing, and lumpy reviews—have well-supported solutions in Anki by tweaking the rules and third-party plugins.)
Duolingo’s half-life regression explicitly models the probability of you recalling a fact as \(2^{-Δ/h}\), where Δ is the time since your last review and \(h\) is a half-life. In this model, your chances of passing a quiz after \(h\) days is 50%, which drops to 25% after \(2 h\) days. They estimate this half-life by combining your past performance and fact metadata in a large-scale machine learning technique called half-life regression (a variant of logistic regression or beta regression, more tuned to this forgetting curve). With each fact associated with a half-life, they can predict the likelihood of forgetting a fact if a quiz was given right now. The results of that quiz (for whichever fact was chosen to review) are used to update that fact’s half-life by re-running the machine learning process with the results from the latest quizzes.
The Mozer group’s algorithms also fit a hierarchical Bayesian model that links quiz performance to memory, taking into account inter-fact and inter-student variability, but the training step is again computationally-intensive.
Like Duolingo and Mozer’s approaches, Ebisu explicitly tracks the exponential forgetting curve to provide a list of facts sorted by most to least likely to be forgotten. However, Ebisu formulates the problem very differently—while memory is understood to decay exponentially, Ebisu posits a probability distribution on the half-life and uses quiz results to update its beliefs in a fully Bayesian way. These updates, while a bit more computationally-burdensome than Anki’s scheduler, are much lighter-weight than Duolingo’s industrial-strength approach.
This gives small quiz apps the same intelligent scheduling as Duolingo’s approach—real-time recall probabilities for any fact—but with immediate incorporation of quiz results, even on mobile apps.
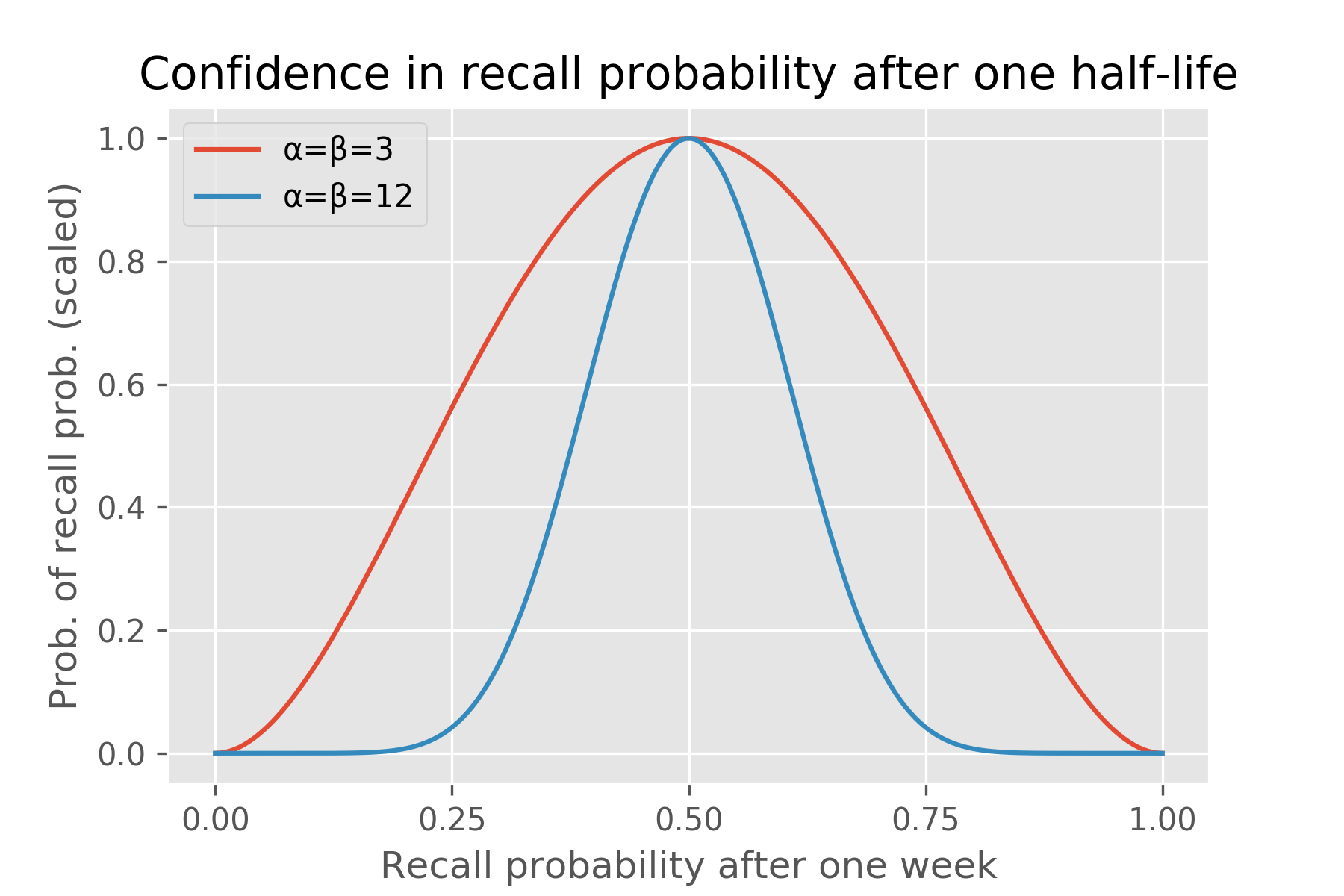
To appreciate this further, consider this example. Imagine a fact with half-life of a week: after a week we expect the recall probability to drop to 50%. However, Ebisu can entertain an infinite range of beliefs about this recall probability: it can be very uncertain that it’ll be 50% (the “α=β=3” model below), or it can be very confident in that prediction (“α=β=12” case):
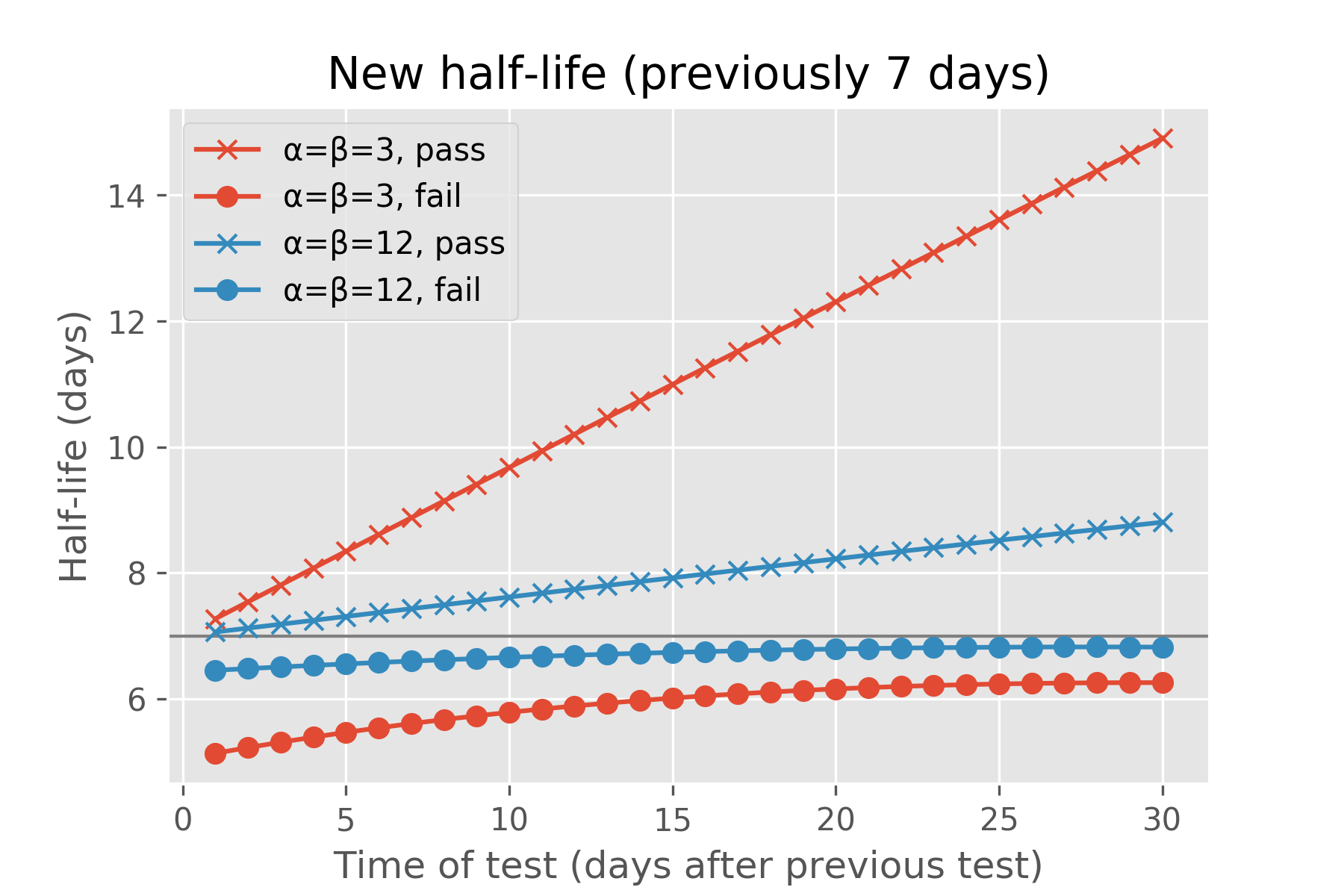
Under either of these models of recall probability, we can ask Ebisu what the expected half-life is after the student is quizzed on this fact a day, a week, or a month after their last review, and whether they passed or failed the quiz:
If the student correctly answers the quiz, Ebisu expects the new half-life to be greater than a week. If the student answers correctly after just a day, the half-life rises a little bit, since we expected the student to remember this fact that soon after reviewing it. If the student surprises us by failing the quiz just a day after they last reviewed it, the projected half-life drops. The more tentative “α=β=3” model aggressively adjusts the half-life, while the more assured “α=β=12” model is more conservative in its update. (Each fact has an α and β associated with it and I explain what they mean mathematically in the next section. Also, the code for these two charts is below.)
Similarly, if the student fails the quiz after a whole month of not reviewing it, this isn’t a surprise—the half-life drops a bit from the initial half-life of a week. If she does surprise us, passing the quiz after a month of not studying it, then Ebisu boosts its expected half-life—by a lot for the “α=β=3” model, less for the “α=β=12” one.
Currently, Ebisu treats each fact as independent, very much like Ebbinghaus’ nonsense syllables: it does not understand how facts are related the way Duolingo can with its sentences. However, Ebisu can be used in combination with other techniques to accommodate extra information about relationships between facts.
The math
Bernoulli quizzes
Let’s begin with a quiz. One way or another, we’ve picked a fact to quiz the student on, \(t\) days (the units are arbitrary since \(t\) can be any positive real number) after her last quiz on it, or since she learned it for the first time.
We’ll model the results of the quiz as a Bernoulli experiment—we’ll later expand this to a binomial experiment. So for Bernoulli quizzes, \(x_t ∼ Bernoulli(p)\); \(x_t\) can be either 1 (success) with probability \(p_t\), or 0 (fail) with probability \(1-p_t\). Let’s think about \(p_t\) as the recall probability at time \(t\)—then \(x_t\) is a coin flip, with a \(p_t\)-weighted coin.
The Beta distribution happens to be the conjugate prior for the Bernoulli distribution. So if our a priori belief about \(p_t\) follow a Beta distribution, that is, if \[p_t ∼ Beta(α_t, β_t)\] for specific \(α_t\) and \(β_t\), then observing the quiz result updates our belief about the recall probability to be: \[p_t | x_t ∼ Beta(α_t + x_t, β_t + 1 - x_t).\]
Aside 0 If you see a gibberish above instead of a mathematical equation (it can be hard to tell the difference sometimes…), you’re probably reading this on GitHub instead of the main Ebisu website which has typeset all equations with MathJax. Read this document there.
Aside 1 Notice that since \(x_t\) is either 1 or 0, the updated parameters \((α + x_t, β + 1 - x_t)\) are \((α + 1, β)\) when the student correctly answered the quiz, and \((α, β + 1)\) when she answered incorrectly.
Aside 2 Even if you’re familiar with Bayesian statistics, if you’ve never worked with priors on probabilities, the meta-ness here might confuse you. What the above means is that, before we flipped our \(p_t\)-weighted coin (before we administered the quiz), we had a specific probability distribution representing the coin’s weighting \(p_t\), not just a scalar number. After we observed the result of the coin flip, we updated our belief about the coin’s weighting—it still makes total sense to talk about the probability of something happening after it happens. Said another way, since we’re being Bayesian, something actually happening doesn’t preclude us from maintaining beliefs about what could have happened.
This is totally ordinary, bread-and-butter Bayesian statistics. However, the major complication arises when the experiment took place not at time \(t\) but \(t_2\): we had a Beta prior on \(p_t\) (probability of recall at time \(t\)) but the test is administered at some other time \(t_2\).
How can we update our beliefs about the recall probability at time \(t\) to another time \(t_2\), either earlier or later than \(t\)?
Moving Beta distributions through time
Our old friend Ebbinghaus comes to our rescue. According to the exponentially-decaying forgetting curve, the probability of recall at time \(t\) is \[p_t = 2^{-t/h},\] for some notional half-life \(h\). Let \(t_2 = δ·t\). Then, \[p_{t_2} = p_{δ t} = 2^{-δt/h} = (2^{-t/h})^δ = (p_t)^δ.\] That is, to fast-forward or rewind \(p_t\) to time \(t_2\), we raise it to the \(δ = t_2 / t\) power.
Unfortunately, a Beta-distributed \(p_t\) becomes non-Beta-distributed when raised to any positive power \(δ\). For a quiz with recall probability given by \(p_t ∼ Beta(12, 12)\) for \(t\) one week after the last review (the middle histogram below), \(δ > 1\) shifts the density to the left (lower recall probability) while \(δ < 1\) does the opposite. Below shows the histogram of recall probability at the original half-life of seven days compared to that after two days (\(δ = 0.3\)) and three weeks (\(δ = 3\)).
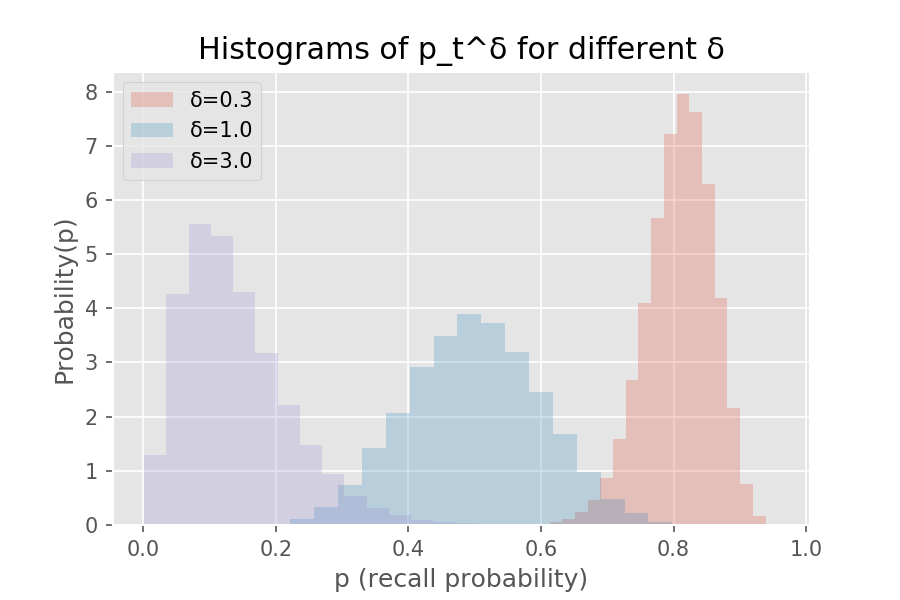
We could approximate this \(δ\) with a Beta random variable, but especially when over- or under-reviewing, the closest Beta fit is very poor. So let’s derive analytically the probability density function (PDF) for \(p_t^δ\). Recall the conventional way to obtain the density of a nonlinearly-transformed random variable: let \(x=p_t\) and \(y = g(x) = x^δ\) be the forward transform, so \(g^{-1}(y) = y^{1/δ}\) is its inverse. Then, with \(P_X(x) = Beta(x; α,β)\), \[P_{Y}(y) = P_{X}(g^{-1}(y)) · \frac{∂}{∂y} g^{-1}(y),\] and this after some Wolfram Alpha and hand-manipulation becomes \[P_{Y}(y) = y^{(α-δ)/δ} · (1-y^{1/δ})^{β-1} / (δ · B(α, β)),\] where \(B(α, β) = Γ(α) · Γ(β) / Γ(α + β)\) is beta function, also the normalizing denominator in the Beta density (confusing, sorry), and \(Γ(·)\) is the gamma function, a generalization of factorial.
To check this, type in
y^((a-1)/d) * (1 - y^(1/d))^(b-1) / Beta[a,b] * D[y^(1/d), y]at Wolfram Alpha.
Replacing the \(X\)’s and \(Y\)’s with our usual variables, we have the probability density for \(p_{t_2} = p_t^δ\) in terms of the original density for \(p_t\): \[P(p_t^δ) = \frac{p^{(α - δ)/δ} · (1-p^{1/δ})^{β-1}}{δ · B(α, β)}.\]
Robert Kern noticed that this is a generalized Beta of the first kind, or GB1, random variable: \[p_t^δ ∼ GB1(p; 1/δ, 1, α; β)\] When \(δ=1\), that is, at exactly the half-life, recall probability is simply the initial Beta we started with.
We will use the density of \(p_t^δ\) to reach our two most important goals:
- what’s the recall probability of a given fact right now?, and
- how do I update my estimate of that recall probability given quiz results?
Recall probability right now
Let’s see how to get the recall probability right now. Recall that we started out with a prior on the recall probabilities \(t\) days after the last review, \(p_t ∼ Beta(α, β)\). Letting \(δ = t_{now} / t\), where \(t_{now}\) is the time currently elapsed since the last review, we saw above that \(p_t^δ\) is GB1-distributed. Wikipedia kindly gives us an expression for the expected recall probability right now, in terms of the Beta function, which we may as well simplify to Gamma function evaluations: \[ E[p_t^δ] = \frac{B(α+δ, β)}{B(α,β)} = \frac{Γ(α + β)}{Γ(α)} · \frac{Γ(α + δ)}{Γ(α + β + δ)} \]
A quiz app can calculate the average current recall probability for each fact using this formula, and thus find the fact most at risk of being forgotten.
Choice of initial model parameters
Mentioning a quiz app reminds me—you may be wondering how to pick the prior triple \([α, β, t]\) initially, for example when the student has first learned a fact.
Set \(t\) equal to your best guess of the fact’s half-life. In Memrise, the first quiz occurs four hours after first learning a fact; in Anki, it’s a day after. To mimic these, set \(t\) to four hours or a day, respectively. In my apps, I set initial \(t\) to a quarter-hour (fifteen minutes).
Then, pick \(α = β > 1\). First, for \(t\) to be a half-life, \(α = β\). Second, a higher value for \(α = β\) means higher confidence that the true half-life is indeed \(t\), which in turn makes the model less sensitive to quiz results—this is, after all, a Bayesian prior. A good default is \(α = β = 3\), which lets the algorithm aggressively change the half-life in response to quiz results.
Quiz apps that allow a students to indicate initial familiarity (or lack thereof) with a flashcard should modify the initial half-life \(t\). It remains an open question whether quiz apps should vary initial \(α = β\) for different flashcards.
Now, let us turn to the final piece of the math, how to update our prior on a fact’s recall probability when a quiz result arrives.
Updating the posterior with quiz results
Recall that our quiz app might ask the student to exercise the same memory multiple times in one sitting, e.g., conjugating the same verb in two different sentences. Therefore, the student’s recall of that memory is a binomial experiment, which is parameterized by \(k\) successes out of \(n\) attempts, with \(0 ≤ k ≤ n\) and \(n ≥ 1\). For many quiz applications, \(n = 1\), so this simplifies to a Bernoulli experiment.
Nota bene. The \(n\) individual sub-trials that make up a single binomial experiment are assumed to be independent of each other. If your quiz application tells the user that, for example, they incorrectly conjugated a verb, and then later in the same review session, asks the user to conjugate the verb again (perhaps in the context of a different sentence), then the two sub-trials are likely not independent, unless the user forgot that they were just asked about that verb. Please get in touch if you want feedback on whether your quiz app design might be running afoul of this caveat.
One option could be this: since we have analytical expressions for the mean and variance of the prior on \(p_t^δ\), convert these to the closest Beta distribution and straightforwardly update with the Bernoulli likelihood as mentioned above, or even the binomial likelihood. However, we can do much better.
By application of Bayes rule, the posterior is \[Posterior(p|k, n) = \frac{Prior(p) · Lik(k|p,n)}{\int_0^1 Prior(p) · Lik(k|p,n) \, dp}.\] Here, “prior” refers to the GB1 density \(P(p_t^δ)\) derived above. \(Lik\) is the binomial likelihood: \(Lik(k|p,n) = \binom{n}{k} p^k (1-p)^{n-k}\). The denominator is the marginal probability of the observation \(k\). (In the above, all recall probabilities \(p\) and quiz results \(k\) are at the same \(t_2 = t · δ\), but we’ll add time subscripts again below.)
Combining all these into one expression, we have: \[ Posterior(p|k, n) = \frac{ p^{α/δ - 1} (1-p^{1/δ})^{β - 1} p^k (1-p)^{n-k} }{ \int_0^1 p^{α/δ - 1} (1-p^{1/δ})^{β - 1} p^k (1-p)^{n-k} \, dp }, \] where note that the big integrand in the denominator is just the numerator.
We use two helpful facts now. The more important one is that \[ \int_0^1 p^{α/δ - 1} (1-p^{1/δ})^{β - 1} \, dp = δ ⋅ B(α, β), \] when \(α, β, δ > 0\). We’ll use this fact several times in what follows—you can see the form of this integrand in the big integrand in the above posterior.
The second helpful fact gets us around that pesky \((1-p)^{n-k}\). By applying the binomial theorem, we can see that \[ \int_0^1 f(x) (1-x)^n \, dx = \sum_{i=0}^{n} \left[ \binom{n}{i} (-1)^i \int_0^1 x^i f(x) \, dx \right], \] for integer \(n > 0\).
Putting these two facts to use, we can show that the posterior at time \(t_2\) is \[ Posterior(p|k, n) = \frac{ \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i p^{α / δ + k + i - 1} (1-p^{1/δ})^{β - 1} }{ δ \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i ⋅ B(α + δ (k + i), \, β) }. \]
This is the posterior at time \(t_2\), the time of the quiz. I’d like to have a posterior at any arbitrary time \(t'\), just in case \(t_2\) happens to be very small or very large. It turns out this posterior can be analytically time-transformed just like we did in the Moving Beta distributions through time section above, except instead of moving a Beta through time, we move this analytic posterior. Just as we have \(δ=t_2/t\) to go from \(t\) to \(t_2\), let \(ε=t' / t_2\) to go from \(t_2\) to \(t'\).
Then, \(P(p_{t'} | k_{t_2}, n_{t_2}) = Posterior(p^{1/ε}|k_{t_2}, n_{t_2}) ⋅ \frac{1}{ε} p^{1/ε - 1}\): \[ P(p_{t'} | k_{t_2}, n_{t_2}) = \frac{ \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i p^(\frac{α + δ (k + i)}{δ ε} - 1) (1-p^{1/(δε)})^{β - 1} }{ δε \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i ⋅ B(α + δ (k + i), \, β) }. \] The denominator is the same in this \(t'\)-time-shifted posterior since it’s just a normalizing constant (and not a function of probability \(p\)) but the numerator retains the same shape as the original, allowing us to use one of our helpful facts above to derive this transformed posterior’s moments. The \(N\)th moment, \(E[p_{t'}^N] \), is: \[ m_N = \frac{ \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i ⋅ B(α + (i+k)δ + N δ ε, \, β) }{ \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i ⋅ B(α + (i+k)δ, \, β) }. \] With these moments of our final posterior at arbitrary time \(t'\) in hand, we can moment-match to recover a Beta-distributed random variable that serves as the new prior. Recall that a distribution with mean \(μ\) and variance \(σ^2\) can be fit to a Beta distribution with parameters:
- \(\hat α = (μ(1-μ)/σ^2 - 1) ⋅ μ\) and
- \(\hat β = (μ(1-μ)/σ^2 - 1) ⋅ (1-μ)\).
In the simple \(n=1\) case of Bernoulli quizzes, these moments simplify further (though in my experience, the code is simpler for the general binominal case).
To summarize the update step: you started with a flashcard whose memory model was \([α, β, t]\). That is, the prior on recall probability after \(t\) time units since the previous encounter is \(Beta(α, β)\). At time \(t_2\), you administer a quiz session that results in \(k\) successful recollections of this flashcard, out of a total of \(n\).
- The updated model is
- \([μ (μ(1-μ)/σ^2 - 1), \, (1-μ) (μ(1-μ)/σ^2 - 1), \, t']\) for any arbitrary time \(t'\), and for
- \(δ = t_2/t\),
- \(ε=t'/t_2\), where both
- \(μ = m_1\) and
- \(σ^2 = m_2 - μ^2\) come from evaluating the appropriate \(m_N\):
- \( m_N = \frac{ \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i ⋅ B(α + (i+k)δ + N δ ε, \, β) }{ \sum_{i=0}^{n-k} \binom{n-k}{i} (-1)^i ⋅ B(α + (i+k)δ, \, β) } \).
- \([μ (μ(1-μ)/σ^2 - 1), \, (1-μ) (μ(1-μ)/σ^2 - 1), \, t']\) for any arbitrary time \(t'\), and for
That’s it! That’s all the math.
Note The Beta function \(B(a,b)=Γ(a) Γ(b) / \Gamma(a+b)\), being a function of a rapidly-growing function like the Gamma function (it is a generalization of factorial), may lose precision in the above expressions for unusual α and β and δ and ε. Addition and subtraction are risky when dealing with floating point numbers that have lost much of their precision. Ebisu takes care to use log-Beta and
logsumexpto minimize loss of precision.
Source code
Before presenting the source code, I must somewhat apologetically explain a bit more about my workflow in writing and editing this document. I use the Atom text editor with the Hydrogen plugin, which allows Atom to communicate with Jupyter kernels. Jupyter used to be called IPython, and is a standard protocol for programming REPLs to communicate with more modern applications like browsers or text editors. With this setup, I can write code in Atom and send it to a behind-the-scenes Python or Node.js or Haskell or Matlab REPL for evaluation, which sends back the result.
Hydrogen developer Lukas Geiger recently added support for evaluating fenced code blocks in Markdown—a long-time dream of mine. This document is a Github-Flavored Markdown file to which I add fenced code blocks. Some of these code blocks I intend to just be demo code, and not end up in the Ebisu library proper, while the code below does need to go into .py files.
In order to untangle the code from the Markdown file to runnable files, I wrote a completely ad hoc undocumented Node.js script called md2code.js which
- slurps the Markdown,
- looks for fenced code blocks that open with a comment indicating a file destination, e.g.,
# export target.py, - prettifies Python with Yapf, JavaScript with clang-format, etc.,
- dumps the code block contents into these files (appending after the first code block), and finally,
- updates the Markdown file itself with this prettified code.
All this enables me to stay in Atom, writing prose and editing/testing code by evaluating fenced code blocks, while also spitting out a proper Python or JavaScript library.
Core library
Python Ebisu contains a sub-module called ebisu.alternate which contains a number of alternative implementations of predictRecall and updateRecall. The __init__ file sets up this module hierarchy.
# export ebisu/__init__.py #
from .ebisu import *
from . import alternate
The above is in its own fenced code block because I don’t want Hydrogen to evaluate it. In Atom, I don’t work with the Ebisu module—I just interact with the raw functions.
Let’s present our Python implementation of the core Ebisu functions, predictRecall and updateRecall, and a couple of other related functions that live in the main ebisu module. All these functions consume a model encoding a Beta prior on recall probabilities at time \(t\), consisting of a 3-tuple containing \((α, β, t)\). I could have gone all object-oriented here but I chose to leave all these functions as stand-alone functions that consume and transform this 3-tuple because (1) I’m not an OOP devotee, and (2) I wanted to maximize the transparency of of this implementation so it can readily be ported to non-OOP, non-Pythonic languages.
Important Note how none of these functions deal with timestamps. All time is captured in “time since last review”, and your external application has to assign units and store timestamps (as illustrated in the Ebisu Jupyter Notebook). This is a deliberate choice! Ebisu wants to know as little about your facts as possible.
In the math section above we derived the mean recall probability at time \(t_2 = t · δ\) given a model \(α, β, t\): \(E[p_t^δ] = B(α+δ, β)/B(α,β)\), which is readily computed using Scipy’s log-beta to avoid overflowing and precision-loss in predictRecall (🍏 below).
As a computational speedup, we can skip the final exp that converts the probability from the log-domain to the linear domain as long as we don’t need an actual probability (i.e., a number between 0 and 1). The output of the function will then be a “pseudo-probability” and can be compared to other “pseudo-probabilities” are returned by the function to rank forgetfulness. Taking advantage of this can, for one example, reduce the runtime from 5.69 µs (± 158 ns) to 4.01 µs (± 215 ns), a 1.4× speedup.
Another computational speedup is that we can cache calls to \(B(α,β)\), which don’t change when the function is called for same quiz repeatedly, as might happen if a quiz app repeatedly asks for the latest recall probability for its flashcards. When the cache is hit, the number of calls to betaln drops from two to one. (Python 3.2 got the nice functools.lru_cache decorator but we forego its use for backwards-compatibility with Python 2.)
# export ebisu/ebisu.py #
from scipy.special import betaln, logsumexp
import numpy as np
def predictRecall(prior, tnow, exact=False):
"""Expected recall probability now, given a prior distribution on it. 🍏
`prior` is a tuple representing the prior distribution on recall probability
after a specific unit of time has elapsed since this fact's last review.
Specifically, it's a 3-tuple, `(alpha, beta, t)` where `alpha` and `beta`
parameterize a Beta distribution that is the prior on recall probability at
time `t`.
`tnow` is the *actual* time elapsed since this fact's most recent review.
Optional keyword parameter `exact` makes the return value a probability,
specifically, the expected recall probability `tnow` after the last review: a
number between 0 and 1. If `exact` is false (the default), some calculations
are skipped and the return value won't be a probability, but can still be
compared against other values returned by this function. That is, if
> predictRecall(prior1, tnow1, exact=True) < predictRecall(prior2, tnow2, exact=True)
then it is guaranteed that
> predictRecall(prior1, tnow1, exact=False) < predictRecall(prior2, tnow2, exact=False)
The default is set to false for computational efficiency.
See README for derivation.
"""
from numpy import exp
a, b, t = prior
dt = tnow / t
ret = betaln(a + dt, b) - _cachedBetaln(a, b)
return exp(ret) if exact else ret
_BETALNCACHE = {}
def _cachedBetaln(a, b):
"Caches `betaln(a, b)` calls in the `_BETALNCACHE` dictionary."
if (a, b) in _BETALNCACHE:
return _BETALNCACHE[(a, b)]
x = betaln(a, b)
_BETALNCACHE[(a, b)] = x
return x
Next is the implementation of updateRecall (🍌 below), which accepts
- a
model(as above, represents the Beta prior on recall probability at one specific time since the fact’s last review), -
successes: the number of times the student successfully exercised this memory, out of -
totaltrials, and -
tnow, the actual time since last quiz that this quiz was administered,
and returns a new model, representing an updated Beta prior on recall probability over some new time horizon. The function implements the update equations above, with an extra rebalancing stage at the end: if the updated α and β are unbalanced (meaning one is much larger than the other), find the half-life of the proposed update and rerun the update for that half-life. At the half-life, the two parameters of the Beta distribution, α and β, will be equal. (To save a few computations, the half-life is calculated via a coarse search, so the rebalanced α and β will likely not be exactly equal.) To facilitate this final rebalancing step, two additional keyword arguments are needed: the time horizon for the update tback, and a rebalance flag to forbid more than one level of rebalancing, and all rebalancing is done in a _rebalance helper function.
(The half-life-finding function is described in more detail below.)
The function uses logsumexp, which seeks to mitigate loss of precision when subtract in the log-domain. A helper function finds the Beta distribution that best matches a given mean and variance, _meanVarToBeta. Another helper function, binomln, computes the logarithm of the binomial expansion, which Scipy does not provide.
# export ebisu/ebisu.py #
def binomln(n, k):
"Log of scipy.special.binom calculated entirely in the log domain"
return -betaln(1 + n - k, 1 + k) - np.log(n + 1)
def updateRecall(prior, successes, total, tnow, rebalance=True, tback=None):
"""Update a prior on recall probability with a quiz result and time. 🍌
`prior` is same as in `ebisu.predictRecall`'s arguments: an object
representing a prior distribution on recall probability at some specific time
after a fact's most recent review.
`successes` is the number of times the user *successfully* exercised this
memory during this review session, out of `total` attempts. Therefore, `0 <=
successes <= total` and `1 <= total`.
If the user was shown this flashcard only once during this review session,
then `total=1`. If the quiz was a success, then `successes=1`, else
`successes=0`.
If the user was shown this flashcard *multiple* times during the review
session (e.g., Duolingo-style), then `total` can be greater than 1.
`tnow` is the time elapsed between this fact's last review and the review
being used to update.
(The keyword arguments `rebalance` and `tback` are intended for internal use.)
Returns a new object (like `prior`) describing the posterior distribution of
recall probability at `tback` (which is an optional input, defaults to
`tnow`).
N.B. This function is tested for numerical stability for small `total < 5`. It
may be unstable for much larger `total`.
N.B.2. This function may throw an assertion error upon numerical instability.
This can happen if the algorithm is *extremely* surprised by a result; for
example, if `successes=0` and `total=5` (complete failure) when `tnow` is very
small compared to the halflife encoded in `prior`. Calling functions are asked
to call this inside a try-except block and to handle any possible
`AssertionError`s in a manner consistent with user expectations, for example,
by faking a more reasonable `tnow`. Please open an issue if you encounter such
exceptions for cases that you think are reasonable.
"""
assert (0 <= successes and successes <= total and 1 <= total)
(alpha, beta, t) = prior
if tback is None:
tback = t
dt = tnow / t
et = tback / tnow
binomlns = [binomln(total - successes, i) for i in range(total - successes + 1)]
logDenominator, logMeanNum, logM2Num = [
logsumexp([
binomlns[i] + betaln(beta, alpha + dt * (successes + i) + m * dt * et)
for i in range(total - successes + 1)
],
b=[(-1)**i
for i in range(total - successes + 1)])
for m in range(3)
]
mean = np.exp(logMeanNum - logDenominator)
m2 = np.exp(logM2Num - logDenominator)
message = dict(
prior=prior, successes=successes, total=total, tnow=tnow, rebalance=rebalance, tback=tback)
assert mean > 0, message
assert m2 > 0, message
meanSq = np.exp(2 * (logMeanNum - logDenominator))
var = m2 - meanSq
assert var > 0, message
newAlpha, newBeta = _meanVarToBeta(mean, var)
proposed = newAlpha, newBeta, tback
return _rebalace(prior, successes, total, tnow, proposed) if rebalance else proposed
def _rebalace(prior, k, n, tnow, proposed):
newAlpha, newBeta, _ = proposed
if (newAlpha > 2 * newBeta or newBeta > 2 * newAlpha):
roughHalflife = modelToPercentileDecay(proposed, coarse=True)
return updateRecall(prior, k, n, tnow, rebalance=False, tback=roughHalflife)
return proposed
def _meanVarToBeta(mean, var):
"""Fit a Beta distribution to a mean and variance."""
# [betaFit] https://en.wikipedia.org/w/index.php?title=Beta_distribution&oldid=774237683#Two_unknown_parameters
tmp = mean * (1 - mean) / var - 1
alpha = mean * tmp
beta = (1 - mean) * tmp
return alpha, beta
Finally we have a couple more helper functions in the main ebisu namespace.
Although our update function above explicitly computes an approximate-half-life for a memory model, it may be very useful to predict when a given memory model expects recall to decay to an arbitrary percentile, not just 50% (i.e., half-life). Besides feedback to users, a quiz app might store the time when each quiz’s recall probability reaches 50%, 5%, 0.05%, …, as a computationally-efficient approximation to the exact recall probability. I am grateful to Robert Kern for contributing the modelToPercentileDecay function (🏀 below). It takes a model, and optionally a percentile keyword (a number between 0 and 1), as well as a coarse flag. The full half-life search does a coarse grid search and then refines that result with numerical optimization. When coarse=True (as in the updateRecall function above), the final finishing optimization is skipped.
The least important function from a usage point of view is also the most important function for someone getting started with Ebisu: I call it defaultModel (🍗 below) and it simply creates a “model” object (a 3-tuple) out of the arguments it’s given. It’s included in the ebisu namespace to help developers who totally lack confidence in picking parameters: the only information it absolutely needs is an expected half-life, e.g., four hours or twenty-four hours or however long you expect a newly-learned fact takes to decay to 50% recall.
# export ebisu/ebisu.py #
def modelToPercentileDecay(model, percentile=0.5, coarse=False):
"""When will memory decay to a given percentile? 🏀
Given a memory `model` of the kind consumed by `predictRecall`,
etc., and optionally a `percentile` (defaults to 0.5, the
half-life), find the time it takes for memory to decay to
`percentile`. If `coarse`, the returned time (in the same units as
`model`) is approximate.
"""
# Use a root-finding routine in log-delta space to find the delta that
# will cause the GB1 distribution to have a mean of the requested quantile.
# Because we are using well-behaved normalized deltas instead of times, and
# owing to the monotonicity of the expectation with respect to delta, we can
# quickly scan for a rough estimate of the scale of delta, then do a finishing
# optimization to get the right value.
assert (percentile > 0 and percentile < 1)
from scipy.special import betaln
from scipy.optimize import root_scalar
alpha, beta, t0 = model
logBab = betaln(alpha, beta)
logPercentile = np.log(percentile)
def f(lndelta):
logMean = betaln(alpha + np.exp(lndelta), beta) - logBab
return logMean - logPercentile
# Scan for a bracket.
bracket_width = 1.0 if coarse else 6.0
blow = -bracket_width / 2.0
bhigh = bracket_width / 2.0
flow = f(blow)
fhigh = f(bhigh)
while flow > 0 and fhigh > 0:
# Move the bracket up.
blow = bhigh
flow = fhigh
bhigh += bracket_width
fhigh = f(bhigh)
while flow < 0 and fhigh < 0:
# Move the bracket down.
bhigh = blow
fhigh = flow
blow -= bracket_width
flow = f(blow)
assert flow > 0 and fhigh < 0
if coarse:
return (np.exp(blow) + np.exp(bhigh)) / 2 * t0
sol = root_scalar(f, bracket=[blow, bhigh])
t1 = np.exp(sol.root) * t0
return t1
def defaultModel(t, alpha=3.0, beta=None):
"""Convert recall probability prior's raw parameters into a model object. 🍗
`t` is your guess as to the half-life of any given fact, in units that you
must be consistent with throughout your use of Ebisu.
`alpha` and `beta` are the parameters of the Beta distribution that describe
your beliefs about the recall probability of a fact `t` time units after that
fact has been studied/reviewed/quizzed. If they are the same, `t` is a true
half-life, and this is a recommended way to create a default model for all
newly-learned facts. If `beta` is omitted, it is taken to be the same as
`alpha`.
"""
return (alpha, beta or alpha, t)
I would expect all the functions above to be present in all implementations of Ebisu:
-
predictRecall, aided by a private helper function_cachedBetaln, -
updateRecall, aided by private helper functions_rebalaceand_meanVarToBeta, -
modelToPercentileDecay, and -
defaultModel.
The functions in the following section are either for illustrative or debugging purposes.
Miscellaneous functions
I wrote a number of other functions that help provide insight or help debug the above functions in the main ebisu workspace but are not necessary for an actual implementation. These are in the ebisu.alternate submodule and not nearly as much time has been spent on polish or optimization as the above core functions. However they are very helpful in unit tests.
# export ebisu/alternate.py #
from .ebisu import _meanVarToBeta
predictRecallMode and predictRecallMedian return the mode and median of the recall probability prior rewound or fast-forwarded to the current time. That is, they return the mode/median of the random variable \(p_t^δ\) whose mean is returned by predictRecall (🍏 above). Recall that \(δ = t / t_{now}\).
Both median and mode, like the mean, have analytical expressions. The mode is a little dangerous: the distribution can blow up to infinity at 0 or 1 when \(δ\) is either much smaller or much larger than 1, in which case the analytical expression for mode may yield nonsense—I have a number of not-very-rigorous checks to attempt to detect this. The median is computed with a inverse incomplete Beta function (betaincinv), and could replace the mean as predictRecall’s return value in a future version of Ebisu.
predictRecallMonteCarlo is the simplest function. It evaluates the mean, variance, mode (via histogram), and median of \(p_t^δ\) by drawing samples from the Beta prior on \(p_t\) and raising them to the \(δ\)-power. The unit tests for predictRecall in the next section use this Monte Carlo to test both derivations and implementations. While fool-proof, Monte Carlo simulation is obviously far too computationally-burdensome for regular use.
# export ebisu/alternate.py #
import numpy as np
def _logsubexp(a, b):
"""Evaluate `log(exp(a) - exp(b))` preserving accuracy.
Subtract log-domain numbers and return in the log-domain.
Wraps `scipy.special.logsumexp`.
"""
from scipy.special import logsumexp
return logsumexp([a, b], b=[1, -1])
def predictRecallMode(prior, tnow):
"""Mode of the immediate recall probability.
Same arguments as `ebisu.predictRecall`, see that docstring for details. A
returned value of 0 or 1 may indicate divergence.
"""
# [1] Mathematica: `Solve[ D[p**((a-t)/t) * (1-p**(1/t))**(b-1), p] == 0, p]`
alpha, beta, t = prior
dt = tnow / t
pr = lambda p: p**((alpha - dt) / dt) * (1 - p**(1 / dt))**(beta - 1)
# See [1]. The actual mode is `modeBase ** dt`, but since `modeBase` might
# be negative or otherwise invalid, check it.
modeBase = (alpha - dt) / (alpha + beta - dt - 1)
if modeBase >= 0 and modeBase <= 1:
# Still need to confirm this is not a minimum (anti-mode). Do this with a
# coarse check of other points likely to be the mode.
mode = modeBase**dt
modePr = pr(mode)
eps = 1e-3
others = [
eps, mode - eps if mode > eps else mode / 2, mode + eps if mode < 1 - eps else
(1 + mode) / 2, 1 - eps
]
otherPr = map(pr, others)
if max(otherPr) <= modePr:
return mode
# If anti-mode detected, that means one of the edges is the mode, likely
# caused by a very large or very small `dt`. Just use `dt` to guess which
# extreme it was pushed to. If `dt` == 1.0, and we get to this point, likely
# we have malformed alpha/beta (i.e., <1)
return 0.5 if dt == 1. else (0. if dt > 1 else 1.)
def predictRecallMedian(prior, tnow, percentile=0.5):
"""Median (or percentile) of the immediate recall probability.
Same arguments as `ebisu.predictRecall`, see that docstring for details.
An extra keyword argument, `percentile`, is a float between 0 and 1, and
specifies the percentile rather than 50% (median).
"""
# [1] `Integrate[p**((a-t)/t) * (1-p**(1/t))**(b-1) / t / Beta[a,b], p]`
# and see "Alternate form assuming a, b, p, and t are positive".
from scipy.special import betaincinv
alpha, beta, t = prior
dt = tnow / t
return betaincinv(alpha, beta, percentile)**dt
def predictRecallMonteCarlo(prior, tnow, N=1000 * 1000):
"""Monte Carlo simulation of the immediate recall probability.
Same arguments as `ebisu.predictRecall`, see that docstring for details. An
extra keyword argument, `N`, specifies the number of samples to draw.
This function returns a dict containing the mean, variance, median, and mode
of the current recall probability.
"""
import scipy.stats as stats
alpha, beta, t = prior
tPrior = stats.beta.rvs(alpha, beta, size=N)
tnowPrior = tPrior**(tnow / t)
freqs, bins = np.histogram(tnowPrior, 'auto')
bincenters = bins[:-1] + np.diff(bins) / 2
return dict(
mean=np.mean(tnowPrior),
median=np.median(tnowPrior),
mode=bincenters[freqs.argmax()],
var=np.var(tnowPrior))
Next we have a Monte Carlo approach to updateRecall (🍌 above), the deceptively-simple updateRecallMonteCarlo. Like predictRecallMonteCarlo above, it draws samples from the Beta distribution in model and propagates them through Ebbinghaus’ forgetting curve to the time specified. To model the likelihood update from the quiz result, it assigns weights to each sample—each weight is that sample’s probability according to the binomial likelihood. (This is equivalent to multiplying the prior with the likelihood—and we needn’t bother with the marginal because it’s just a normalizing factor which would scale all weights equally. I am grateful to mxwsn for suggesting this elegant approach.) It then applies Ebbinghaus again to move the distribution to tback. Finally, the ensemble is collapsed to a weighted mean and variance to be converted to a Beta distribution.
# export ebisu/alternate.py #
def updateRecallMonteCarlo(prior, k, n, tnow, tback=None, N=10 * 1000 * 1000):
"""Update recall probability with quiz result via Monte Carlo simulation.
Same arguments as `ebisu.updateRecall`, see that docstring for details.
An extra keyword argument `N` specifies the number of samples to draw.
"""
# [bernoulliLikelihood] https://en.wikipedia.org/w/index.php?title=Bernoulli_distribution&oldid=769806318#Properties_of_the_Bernoulli_Distribution, third equation
# [weightedMean] https://en.wikipedia.org/w/index.php?title=Weighted_arithmetic_mean&oldid=770608018#Mathematical_definition
# [weightedVar] https://en.wikipedia.org/w/index.php?title=Weighted_arithmetic_mean&oldid=770608018#Weighted_sample_variance
import scipy.stats as stats
from scipy.special import binom
if tback is None:
tback = tnow
alpha, beta, t = prior
tPrior = stats.beta.rvs(alpha, beta, size=N)
tnowPrior = tPrior**(tnow / t)
# This is the Bernoulli likelihood [bernoulliLikelihood]
weights = binom(n, k) * (tnowPrior)**k * ((1 - tnowPrior)**(n - k))
# Now propagate this posterior to the tback
tbackPrior = tPrior**(tback / t)
# See [weightedMean]
weightedMean = np.sum(weights * tbackPrior) / np.sum(weights)
# See [weightedVar]
weightedVar = np.sum(weights * (tbackPrior - weightedMean)**2) / np.sum(weights)
newAlpha, newBeta = _meanVarToBeta(weightedMean, weightedVar)
return newAlpha, newBeta, tback
That’s it—that’s all the code in the ebisu module!
Test code
I use the built-in unittest, and I can run all the tests from Atom via Hydrogen/Jupyter but for historic reasons I don’t want Jupyter to deal with the ebisu namespace, just functions (since most of these functions and tests existed before the module’s layout was decided). So the following is in its own fenced code block that I don’t evaluate in Atom.
# export ebisu/tests/test_ebisu.py
from ebisu import *
from ebisu.alternate import *
In these unit tests, I compare
-
predictRecallagainstpredictRecallMonteCarlo, and -
updateRecallagainstupdateRecallMonteCarlo.
I also want to make sure that predictRecall and updateRecall both produce sane values when extremely under- and over-reviewing (i.e., immediately after review as well as far into the future) and for a range of successes and total reviews per quiz session. And we should also exercise modelToPercentileDecay.
For testing updateRecall, since all functions return a Beta distribution, I compare the resulting distributions in terms of Kullback–Leibler divergence (actually, the symmetric distance version), which is a nice way to measure the difference between two probability distributions. There is also a little unit test for my implementation for the KL divergence on Beta distributions.
For testing predictRecall, I compare means using relative error, \(|x-y| / |y|\).
For both sets of functions, a range of \(δ = t_{now} / t\) and both outcomes of quiz results (true and false) are tested to ensure they all produce the same answers.
Often the unit tests fails because the tolerances are a little tight, and the random number generator seed is variable, which leads to errors exceeding thresholds. I actually prefer to see these occasional test failures because it gives me confidence that the thresholds are where I want them to be (if I set the thresholds too loose, and I somehow accidentally greatly improved accuracy, I might never know). However, I realize it can be annoying for automated tests or continuous integration systems, so I am open to fixing a seed and fixing the error threshold for it.
One note: the unit tests update a global database of testpoints being tested, which can be dumped to a JSON file for comparison against other implementations.
# export ebisu/tests/test_ebisu.py
import unittest
import numpy as np
def relerr(dirt, gold):
return abs(dirt - gold) / abs(gold)
def maxrelerr(dirts, golds):
return max(map(relerr, dirts, golds))
def klDivBeta(a, b, a2, b2):
"""Kullback-Leibler divergence between two Beta distributions in nats"""
# Via http://bariskurt.com/kullback-leibler-divergence-between-two-dirichlet-and-beta-distributions/
from scipy.special import gammaln, psi
import numpy as np
left = np.array([a, b])
right = np.array([a2, b2])
return gammaln(sum(left)) - gammaln(sum(right)) - sum(gammaln(left)) + sum(
gammaln(right)) + np.dot(left - right,
psi(left) - psi(sum(left)))
def kl(v, w):
return (klDivBeta(v[0], v[1], w[0], w[1]) + klDivBeta(w[0], w[1], v[0], v[1])) / 2.
testpoints = []
class TestEbisu(unittest.TestCase):
def test_predictRecallMedian(self):
model0 = (4.0, 4.0, 1.0)
model1 = updateRecall(model0, 0, 1, 1.0)
model2 = updateRecall(model1, 1, 1, 0.01)
ts = np.linspace(0.01, 4.0, 81)
qs = (0.05, 0.25, 0.5, 0.75, 0.95)
for t in ts:
for q in qs:
self.assertGreater(predictRecallMedian(model2, t, q), 0)
def test_kl(self):
# See https://en.wikipedia.org/w/index.php?title=Beta_distribution&oldid=774237683#Quantities_of_information_.28entropy.29 for these numbers
self.assertAlmostEqual(klDivBeta(1., 1., 3., 3.), 0.598803, places=5)
self.assertAlmostEqual(klDivBeta(3., 3., 1., 1.), 0.267864, places=5)
def test_prior(self):
"test predictRecall vs predictRecallMonteCarlo"
def inner(a, b, t0):
global testpoints
for t in map(lambda dt: dt * t0, [0.1, .99, 1., 1.01, 5.5]):
mc = predictRecallMonteCarlo((a, b, t0), t, N=100 * 1000)
mean = predictRecall((a, b, t0), t, exact=True)
self.assertLess(relerr(mean, mc['mean']), 5e-2)
testpoints += [['predict', [a, b, t0], [t], dict(mean=mean)]]
inner(3.3, 4.4, 1.)
inner(34.4, 34.4, 1.)
def test_posterior(self):
"Test updateRecall via updateRecallMonteCarlo"
def inner(a, b, t0, dts, n=1):
global testpoints
for t in map(lambda dt: dt * t0, dts):
for k in range(n + 1):
msg = 'a={},b={},t0={},k={},n={},t={}'.format(a, b, t0, k, n, t)
an = updateRecall((a, b, t0), k, n, t)
mc = updateRecallMonteCarlo((a, b, t0), k, n, t, an[2], N=1_000_000 * (1 + k))
self.assertLess(kl(an, mc), 5e-3, msg=msg + ' an={}, mc={}'.format(an, mc))
testpoints += [['update', [a, b, t0], [k, n, t], dict(post=an)]]
inner(3.3, 4.4, 1., [0.1, 1., 9.5], n=5)
inner(34.4, 3.4, 1., [0.1, 1., 5.5, 50.], n=5)
def test_update_then_predict(self):
"Ensure #1 is fixed: prediction after update is monotonic"
future = np.linspace(.01, 1000, 101)
def inner(a, b, t0, dts, n=1):
for t in map(lambda dt: dt * t0, dts):
for k in range(n + 1):
msg = 'a={},b={},t0={},k={},n={},t={}'.format(a, b, t0, k, n, t)
newModel = updateRecall((a, b, t0), k, n, t)
predicted = np.vectorize(lambda tnow: predictRecall(newModel, tnow))(future)
self.assertTrue(
np.all(np.diff(predicted) < 0), msg=msg + ' predicted={}'.format(predicted))
inner(3.3, 4.4, 1., [0.1, 1., 9.5], n=5)
inner(34.4, 3.4, 1., [0.1, 1., 5.5, 50.], n=5)
def test_halflife(self):
"Exercise modelToPercentileDecay"
percentiles = np.linspace(.01, .99, 101)
def inner(a, b, t0, dts):
for t in map(lambda dt: dt * t0, dts):
msg = 'a={},b={},t0={},t={}'.format(a, b, t0, t)
ts = np.vectorize(lambda p: modelToPercentileDecay((a, b, t), p))(percentiles)
self.assertTrue(monotonicDecreasing(ts), msg=msg + ' ts={}'.format(ts))
inner(3.3, 4.4, 1., [0.1, 1., 9.5])
inner(34.4, 3.4, 1., [0.1, 1., 5.5, 50.])
def test_asymptotic(self):
"""Failing quizzes in far future shouldn't modify model when updating.
Passing quizzes right away shouldn't modify model when updating.
"""
def inner(a, b, n=1):
prior = (a, b, 1.0)
hl = modelToPercentileDecay(prior)
ts = np.linspace(.001, 1000, 101)
passhl = np.vectorize(
lambda tnow: modelToPercentileDecay(updateRecall(prior, n, n, tnow, 1.0)))(
ts)
failhl = np.vectorize(
lambda tnow: modelToPercentileDecay(updateRecall(prior, 0, n, tnow, 1.0)))(
ts)
self.assertTrue(monotonicIncreasing(passhl))
self.assertTrue(monotonicIncreasing(failhl))
# Passing should only increase halflife
self.assertTrue(np.all(passhl >= hl * .999))
# Failing should only decrease halflife
self.assertTrue(np.all(failhl <= hl * 1.001))
for a in [2., 20, 200]:
for b in [2., 20, 200]:
inner(a, b, n=1)
def monotonicIncreasing(v):
return np.all(np.diff(v) >= -np.spacing(1.) * 1e8)
def monotonicDecreasing(v):
return np.all(np.diff(v) <= np.spacing(1.) * 1e8)
if __name__ == '__main__':
unittest.TextTestRunner().run(unittest.TestLoader().loadTestsFromModule(TestEbisu()))
with open("test.json", "w") as out:
import json
out.write(json.dumps(testpoints))
That if __name__ == '__main__' is for running the test suite in Atom via Hydrogen/Jupyter. I actually use nose to run the tests, e.g., python3 -m nose (which is wrapped in an npm script: if you look in package.json you’ll see that npm test will run the equivalent of node md2code.js && python3 -m "nose": this Markdown file is untangled into Python source files first, and then nose is invoked).
Demo codes
The code snippets here are intended to demonstrate some Ebisu functionality.
Visualizing half-lives
The first snippet produces the half-life plots shown above, and included below, scroll down.
import scipy.stats as stats
import numpy as np
import matplotlib.pyplot as plt
plt.style.use('ggplot')
plt.rcParams['svg.fonttype'] = 'none'
t0 = 7.
ts = np.arange(1, 301.)
ps = np.linspace(0, 1., 200)
ablist = [3, 12]
plt.close('all')
plt.figure()
[
plt.plot(
ps, stats.beta.pdf(ps, ab, ab) / stats.beta.pdf(.5, ab, ab), label='α=β={}'.format(ab))
for ab in ablist
]
plt.legend(loc=2)
plt.xticks(np.linspace(0, 1, 5))
plt.title('Confidence in recall probability after one half-life')
plt.xlabel('Recall probability after one week')
plt.ylabel('Prob. of recall prob. (scaled)')
plt.savefig('figures/models.svg')
plt.savefig('figures/models.png', dpi=300)
plt.show()
plt.figure()
ax = plt.subplot(111)
plt.axhline(y=t0, linewidth=1, color='0.5')
[
plt.plot(
ts,
list(map(lambda t: modelToPercentileDecay(updateRecall((a, a, t0), xobs, t)), ts)),
marker='x' if xobs == 1 else 'o',
color='C{}'.format(aidx),
label='α=β={}, {}'.format(a, 'pass' if xobs == 1 else 'fail'))
for (aidx, a) in enumerate(ablist)
for xobs in [1, 0]
]
plt.legend(loc=0)
plt.title('New half-life (previously {:0.0f} days)'.format(t0))
plt.xlabel('Time of test (days after previous test)')
plt.ylabel('Half-life (days)')
plt.savefig('figures/halflife.svg')
plt.savefig('figures/halflife.png', dpi=300)
plt.show()
Why we work with random variables
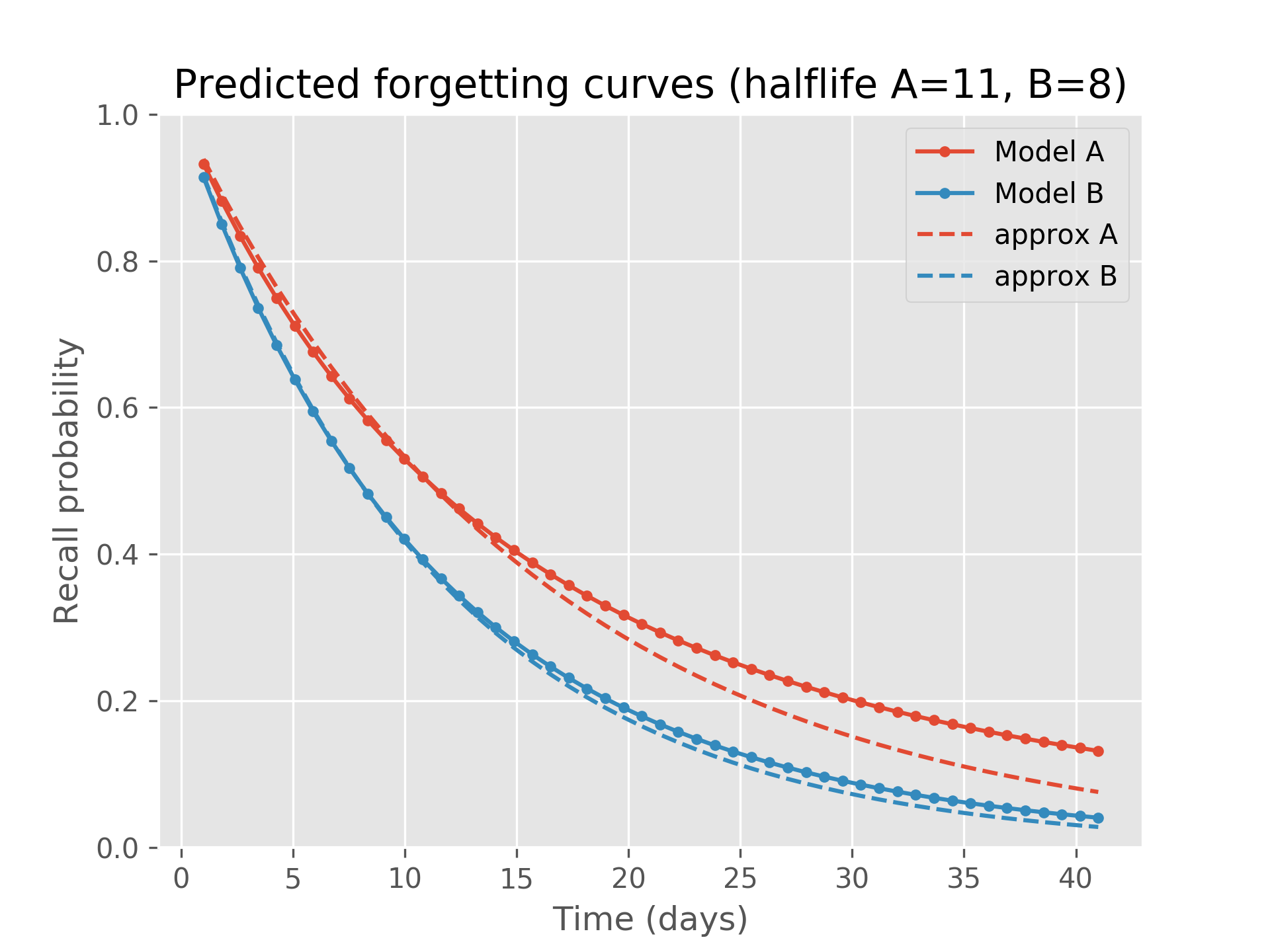
This second snippet addresses a potential approximation which isn’t too accurate but might be useful in some situations. The function predictRecall (🍏 above) in exact mode evaluates the log-gamma function four times and an exp once. One may ask, why not use the half-life returned by modelToPercentileDecay and Ebbinghaus’ forgetting curve, thereby approximating the current recall probability for a fact as 2 ** (-tnow / modelToPercentileDecay(model))? While this is likely more computationally efficient (after computing the half-life up-front), it is also less precise:
ts = np.linspace(1, 41)
modelA = updateRecall((3., 3., 7.), 1, 15.)
modelB = updateRecall((12., 12., 7.), 1, 15.)
hlA = modelToPercentileDecay(modelA)
hlB = modelToPercentileDecay(modelB)
plt.figure()
[
plt.plot(ts, predictRecall(model, ts, exact=True), '.-', label='Model ' + label, color=color)
for model, color, label in [(modelA, 'C0', 'A'), (modelB, 'C1', 'B')]
]
[
plt.plot(ts, 2**(-ts / halflife), '--', label='approx ' + label, color=color)
for halflife, color, label in [(hlA, 'C0', 'A'), (hlB, 'C1', 'B')]
]
# plt.yscale('log')
plt.legend(loc=0)
plt.ylim([0, 1])
plt.xlabel('Time (days)')
plt.ylabel('Recall probability')
plt.title('Predicted forgetting curves (halflife A={:0.0f}, B={:0.0f})'.format(hlA, hlB))
plt.savefig('figures/forgetting-curve.svg')
plt.savefig('figures/forgetting-curve.png', dpi=300)
plt.show()
This plot shows predictRecall’s fully analytical solution for two separate models over time as well as this approximation: model A has half-life of eleven days while model B has half-life of 7.9 days. We see that the approximation diverges a bit from the true solution.
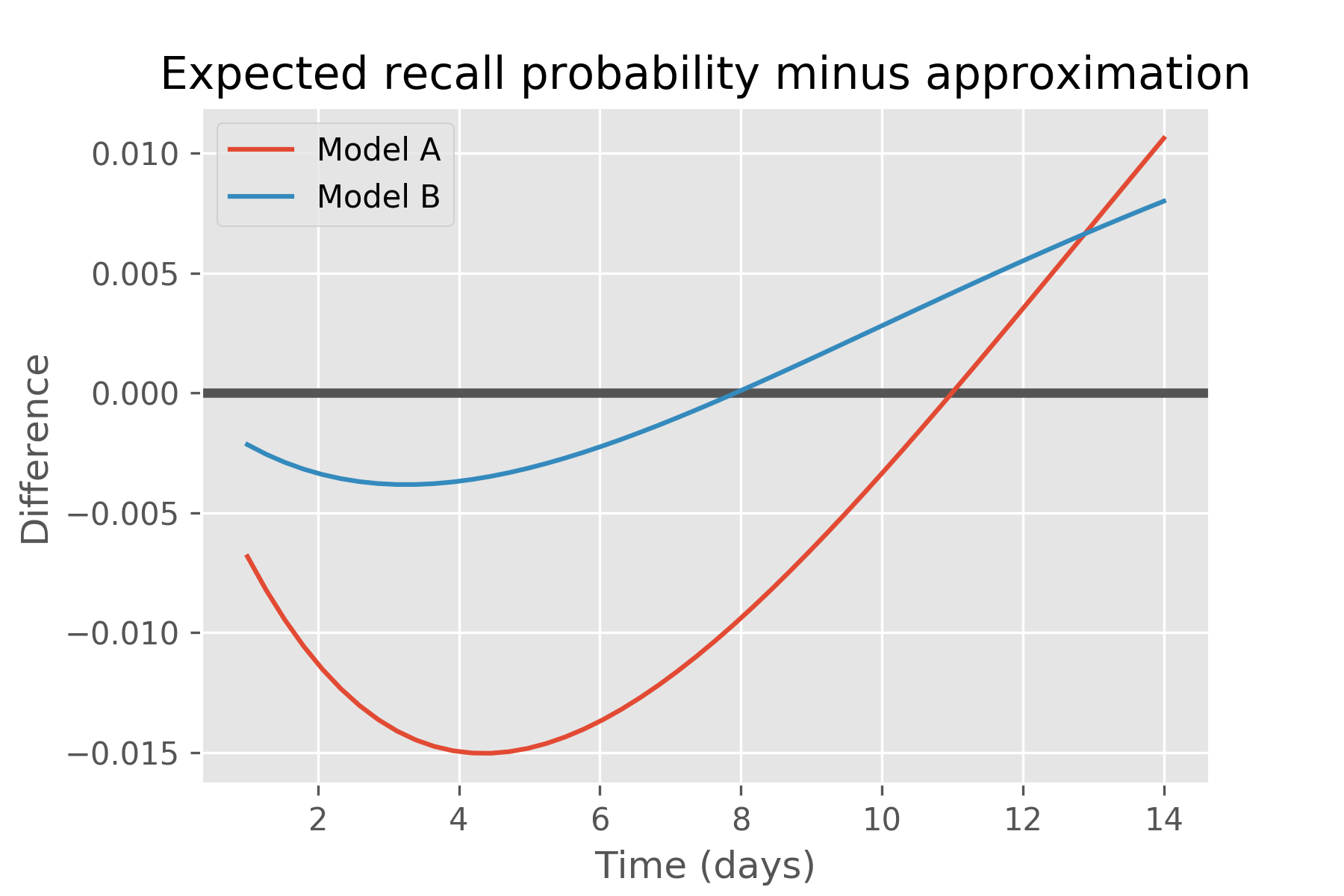
This also indicates that placing a prior on recall probabilities and propagating that prior through time via Ebbinghaus results in a different curve than Ebbinghaus’ exponential decay curve. This surprising result can be seen as a consequence of Jensen’s inequality, which says that \(E[f(p)] ≥ f(E[p])\) when \(f\) is convex, and that the opposite is true if it is concave. In our case, \(f(p) = p^δ\), for δ = t / halflife, and Jensen requires that the accurate mean recall probability is greater than the approximation for times greater than the half-life, and less than otherwise. We see precisely this for both models, as illustrated in this plot of just their differences:
plt.figure()
ts = np.linspace(1, 14)
plt.axhline(y=0, linewidth=3, color='0.33')
plt.plot(ts, predictRecall(modelA, ts, exact=True) - 2**(-ts / hlA), label='Model A')
plt.plot(ts, predictRecall(modelB, ts, exact=True) - 2**(-ts / hlB), label='Model B')
plt.gcf().subplots_adjust(left=0.15)
plt.legend(loc=0)
plt.xlabel('Time (days)')
plt.ylabel('Difference')
plt.title('Expected recall probability minus approximation')
plt.savefig('figures/forgetting-curve-diff.svg')
plt.savefig('figures/forgetting-curve-diff.png', dpi=300)
plt.show()
I think this speaks to the surprising nature of random variables and the benefits of handling them rigorously, as Ebisu seeks to do.
Moving Beta distributions through time
Below is the code to show the histograms on recall probability two days, a week, and three weeks after the last review:
def generatePis(deltaT, alpha=12.0, beta=12.0):
import scipy.stats as stats
piT = stats.beta.rvs(alpha, beta, size=50 * 1000)
piT2 = piT**deltaT
plt.hist(piT2, bins=20, label='δ={}'.format(deltaT), alpha=0.25, normed=True)
[generatePis(p) for p in [0.3, 1., 3.]]
plt.xlabel('p (recall probability)')
plt.ylabel('Probability(p)')
plt.title('Histograms of p_t^δ for different δ')
plt.legend(loc=0)
plt.savefig('figures/pidelta.svg')
plt.savefig('figures/pidelta.png', dpi=150)
plt.show()
Requirements for building all aspects of this repo
- Python
- scipy, numpy
- nose for tests
- Pandoc
- pydoc-markdown
Acknowledgments
A huge thank you to bug reporters and math experts and contributors!
Many thanks to mxwsn and commenters as well as jth for their advice and patience with my statistical incompetence.
Many thanks also to Drew Benedetti for reviewing this manuscript.
John Otander’s Modest CSS is used to style the Markdown output.