IndrajeetPatil / Statsexpressions
Programming Languages
Projects that are alternatives of or similar to Statsexpressions
statsExpressions: Dataframes and expressions with statistical details
| Package | Status | Usage | GitHub | References |
|---|---|---|---|---|
 |
 |
|||
 |
 |
|||
 |
 |
 |
 |
|
 |
 |
|||
 |
 |
 |
||
 |
 |
Introduction 
statsExpressions
produces dataframes with rich details for the most common types of
statistical approaches and tests: parametric, nonparametric, robust, and
Bayesian t-test, one-way ANOVA, correlation analyses, contingency table
analyses, and meta-analyses.
The functions are:
- pipe-friendly
- provide a consistent syntax to work with tidy data
- provide expressions with statistical details for graphing packages
This package forms the statistical processing backend for
ggstatsplot package.
Please note that the API for the package has changed significantly in
1.0.0 release and will break any code that used prior versions of
this package.
See: https://indrajeetpatil.github.io/statsExpressions/news/index.html
Installation
To get the latest, stable CRAN release:
install.packages("statsExpressions")
You can get the development version of the package from GitHub. To
see what new changes (and bug fixes) have been made to the package since
the last release on CRAN, you can check the detailed log of changes
here:
https://indrajeetpatil.github.io/statsExpressions/news/index.html
If you are in hurry and want to reduce the time of installation, prefer-
# needed package to download from GitHub repo
install.packages("remotes")
# downloading the package from GitHub
remotes::install_github(
repo = "IndrajeetPatil/statsExpressions", # package path on GitHub
dependencies = FALSE, # assumes you have already installed needed packages
quick = TRUE # skips docs, demos, and vignettes
)
If time is not a constraint-
remotes::install_github(
repo = "IndrajeetPatil/statsExpressions", # package path on GitHub
dependencies = TRUE, # installs packages which statsExpressions depends on
upgrade_dependencies = TRUE # updates any out of date dependencies
)
Citation
If you want to cite this package in a scientific journal or in any other
context, run the following code in your R console:
citation("statsExpressions")
#>
#> Patil, I. (2018). ggstatsplot: 'ggplot2' Based Plots with Statistical
#> Details. CRAN. Retrieved from
#> https://cran.r-project.org/web/packages/ggstatsplot/index.html
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {{ggstatsplot}: 'ggplot2' Based Plots with Statistical Details},
#> author = {Indrajeet Patil},
#> year = {2018},
#> journal = {CRAN},
#> url = {https://CRAN.R-project.org/package=ggstatsplot},
#> doi = {10.5281/zenodo.2074621},
#> }
Documentation and Examples
To see the documentation relevant for the development version of the
package, see the dedicated website for statsExpressions, which is
updated after every new commit:
https://indrajeetpatil.github.io/statsExpressions/.
Summary of types of statistical analyses
Currently, it supports only the most common types of statistical tests. Specifically, parametric, non-parametric, robust, and bayesian versions of:
- t-test
- anova
- correlation tests
- contingency table analysis
- meta-analysis
The table below summarizes all the different types of analyses currently supported in this package-
| Description | Parametric | Non-parametric | Robust | Bayesian |
|---|---|---|---|---|
| Between group/condition comparisons | Yes | Yes | Yes | Yes |
| Within group/condition comparisons | Yes | Yes | Yes | Yes |
| Distribution of a numeric variable | Yes | Yes | Yes | Yes |
| Correlation between two variables | Yes | Yes | Yes | Yes |
| Association between categorical variables | Yes | NA |
NA |
Yes |
| Equal proportions for categorical variable levels | Yes | NA |
NA |
Yes |
| Random-effects meta-analysis | Yes | NA |
Yes | Yes |
Statistical reporting
For all statistical test expressions, the default template abides by the APA gold standard for statistical reporting. For example, here are results from Yuen’s test for trimmed means (robust t-test):
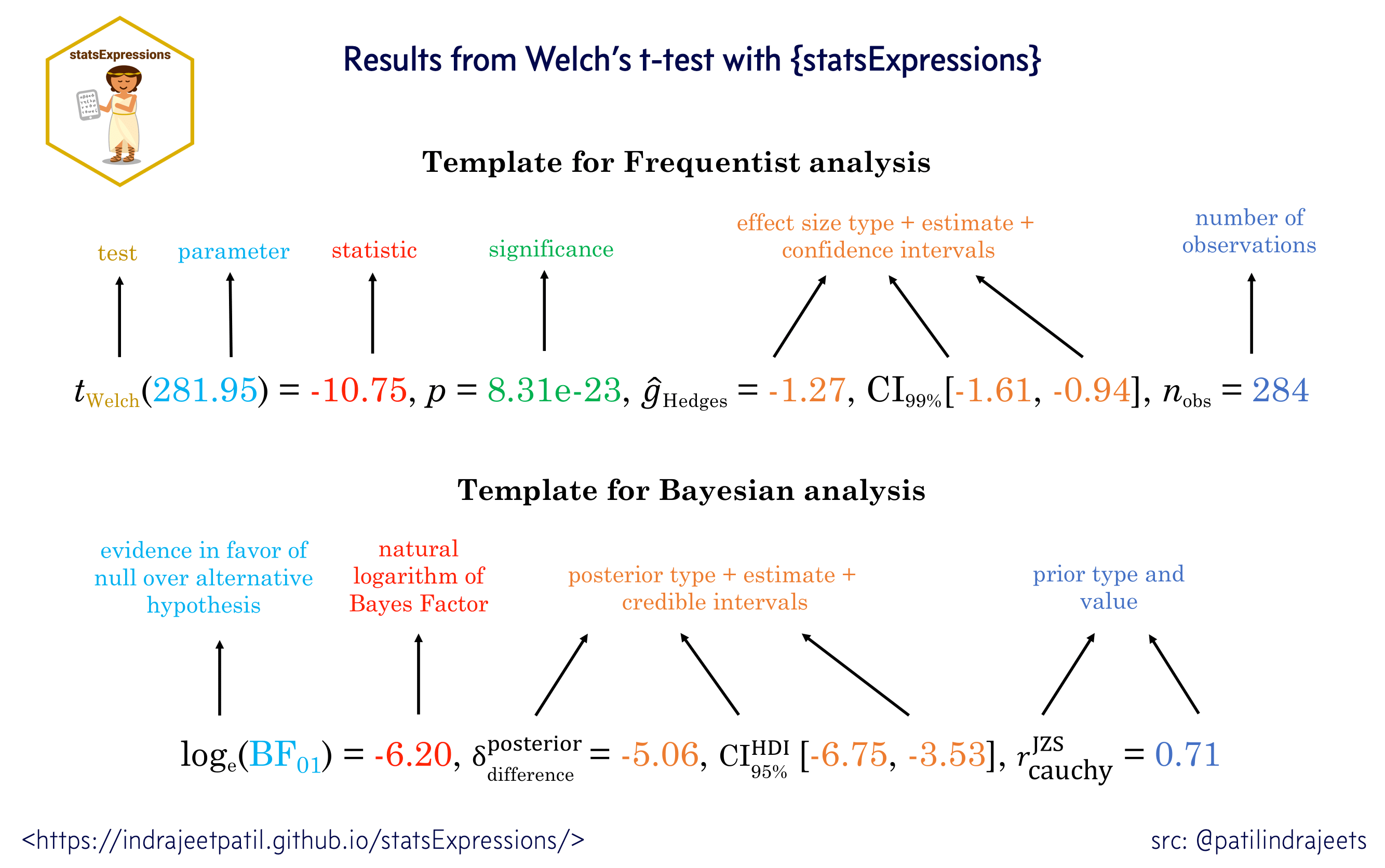
Summary of statistical tests and effect sizes
Here is a summary table of all the statistical tests currently supported across various functions: https://indrajeetpatil.github.io/statsExpressions/articles/stats_details.html
Dataframe as output
The returned dataframe will contain the following columns (the exact columns will depend on the test and the statistical approach):
-
statistic: the numeric value of a statistic.
-
df: the numeric value of a parameter being modeled (often degrees of freedom for the test); note that if
no.parameters = 0L(e.g., for non-parametric tests), this column will be irrelevant. -
df.error, df: relevant only if the statistic in question has two degrees of freedom (e.g., anova).
-
p.value: the two-sided p-value associated with the observed statistic.
-
method: the details of the statistical test carried out.
-
estimate: estimated value of the effect size.
-
conf.low: lower bound for effect size estimate.
-
conf.high: upper bound for effect size estimate.
-
conf.level: width of the confidence interval.
-
effectsize: the details of the effect size.
All possible outputs from all functions are tabulated here: https://indrajeetpatil.github.io/statsExpressions/articles/web_only/dataframe_outputs.html
But here is one quick example:
# setup
library(statsExpressions)
set.seed(123)
# one-way ANOVA - parametric
mtcars %>% oneway_anova(x = cyl, y = wt)
#> # A tibble: 1 x 11
#> statistic df df.error p.value
#> <dbl> <dbl> <dbl> <dbl>
#> 1 20.2 2 19.0 0.0000196
#> method estimate conf.level
#> <chr> <dbl> <dbl>
#> 1 One-way analysis of means (not assuming equal variances) 0.637 0.95
#> conf.low conf.high effectsize expression
#> <dbl> <dbl> <chr> <list>
#> 1 0.309 0.785 Omega2 <language>
Needless to say this will also work with the kable function to
generate a table:
# setup
library(statsExpressions)
set.seed(123)
# one-sample robust t-test
# we will leave `expression` column out; it's not needed for using only the dataframe
mtcars %>%
one_sample_test(wt, test.value = 3, type = "robust") %>%
dplyr::select(-expression) %>%
knitr::kable()
| statistic | p.value | method | estimate | conf.low | conf.high | conf.level | effectsize |
|---|---|---|---|---|---|---|---|
| 1.179181 | 0.22 | Bootstrap-t method for one-sample test | 3.197 | 2.872163 | 3.521837 | 0.95 | Trimmed mean |
These functions also play nicely with dplyr function. For example,
let’s say we want to run a one-sample t-test for all levels of a
certain grouping variable. Here is how you can do it:
# for reproducibility
set.seed(123)
library(dplyr)
# grouped operation
mtcars %>%
group_by(cyl) %>%
group_modify(~ one_sample_test(.x, wt, test.value = 3), .keep = TRUE) %>%
ungroup()
#> # A tibble: 3 x 12
#> cyl mu statistic df.error p.value method estimate conf.level
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 4 3 -4.16 10 0.00195 One Sample t-test -1.16 0.95
#> 2 6 3 0.870 6 0.418 One Sample t-test 0.286 0.95
#> 3 8 3 4.92 13 0.000278 One Sample t-test 1.24 0.95
#> conf.low conf.high effectsize expression
#> <dbl> <dbl> <chr> <list>
#> 1 -1.97 -0.422 Hedges' g <language>
#> 2 -0.419 1.01 Hedges' g <language>
#> 3 0.565 1.98 Hedges' g <language>
Using expressions in custom plots
A list of primary functions in this package can be found at the package website: https://indrajeetpatil.github.io/statsExpressions/reference/index.html
Following are few examples of how these functions can be used.
Example: Expressions for one-way ANOVAs
Between-subjects design
Let’s say we want to check differences in weight of the vehicle based on number of cylinders in the engine and wish to carry out Welch’s ANOVA:
# setup
set.seed(123)
library(ggplot2)
library(ggforce)
library(statsExpressions)
# plot with subtitle
ggplot(iris, aes(x = Species, y = Sepal.Length)) +
geom_violin() +
geom_sina() +
labs(
title = "Fisher's one-way ANOVA",
subtitle = oneway_anova(iris, Species, Sepal.Length, var.equal = TRUE)$expression[[1]]
)

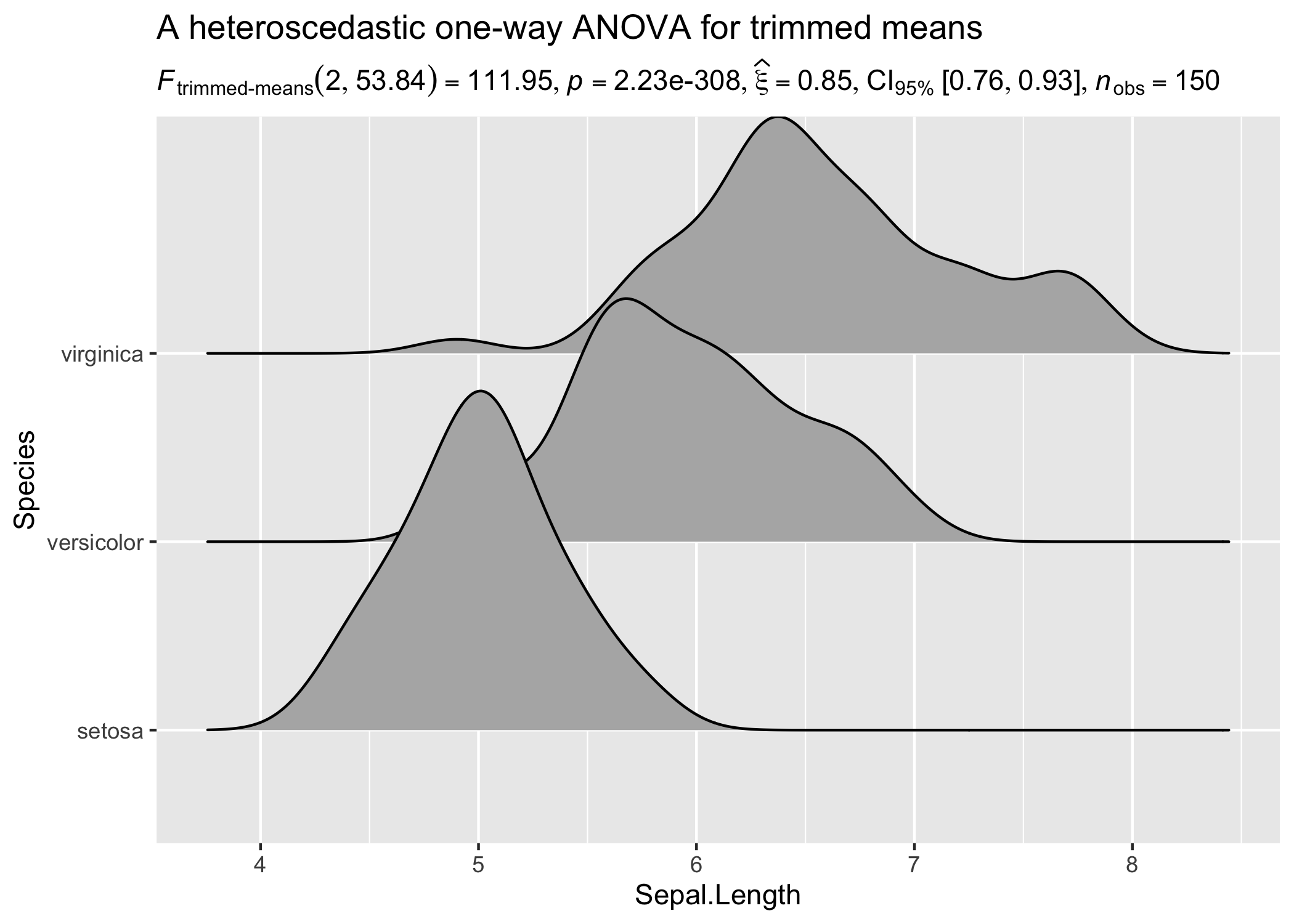
In case you change your mind and now want to carry out a robust ANOVA instead. Also, let’s use a different kind of a visualization:
# setup
set.seed(123)
library(ggplot2)
library(statsExpressions)
library(ggridges)
# create a ridgeplot
ggplot(iris, aes(x = Sepal.Length, y = Species)) +
geom_density_ridges(
jittered_points = TRUE, quantile_lines = TRUE,
scale = 0.9, vline_size = 1, vline_color = "red",
position = position_raincloud(adjust_vlines = TRUE)
) +
labs(
title = "A heteroscedastic one-way ANOVA for trimmed means",
subtitle = oneway_anova(iris, Species, Sepal.Length, type = "robust")$expression[[1]]
)
Needless to say, you can also use these functions to display results in
ggplot-extension packages. For example, ggpubr:
set.seed(123)
library(ggpubr)
library(ggplot2)
# plot
ggboxplot(
ToothGrowth,
x = "dose",
y = "len",
color = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"),
add = "jitter",
shape = "dose"
) + # adding results from stats analysis using `statsExpressions`
labs(
title = "Kruskall-Wallis test",
subtitle = oneway_anova(ToothGrowth, dose, len, type = "np")$expression[[1]]
)

Within-subjects design
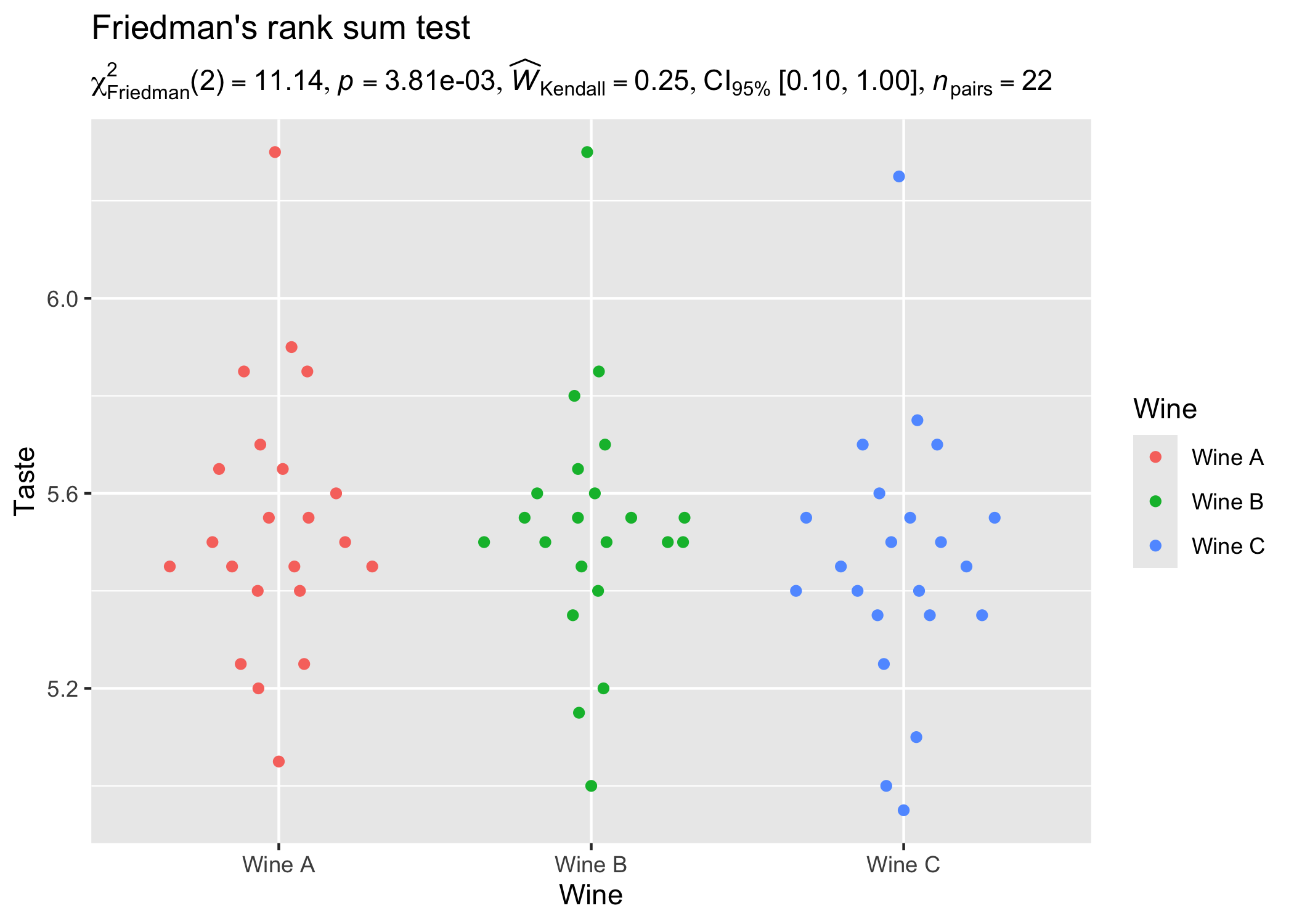
Let’s now see an example of a repeated measures one-way ANOVA.
# setup
set.seed(123)
library(ggplot2)
library(WRS2)
library(ggbeeswarm)
library(statsExpressions)
ggplot2::ggplot(WineTasting, aes(Wine, Taste, color = Wine)) +
geom_quasirandom() +
labs(
title = "Friedman's rank sum test",
subtitle = oneway_anova(
WineTasting,
Wine,
Taste,
paired = TRUE,
subject.id = Taster,
type = "np"
)$expression[[1]]
)
Example: Expressions for two-sample tests
Between-subjects design
# setup
set.seed(123)
library(ggplot2)
library(gghalves)
library(ggbeeswarm)
library(hrbrthemes)
library(statsExpressions)
# create a plot
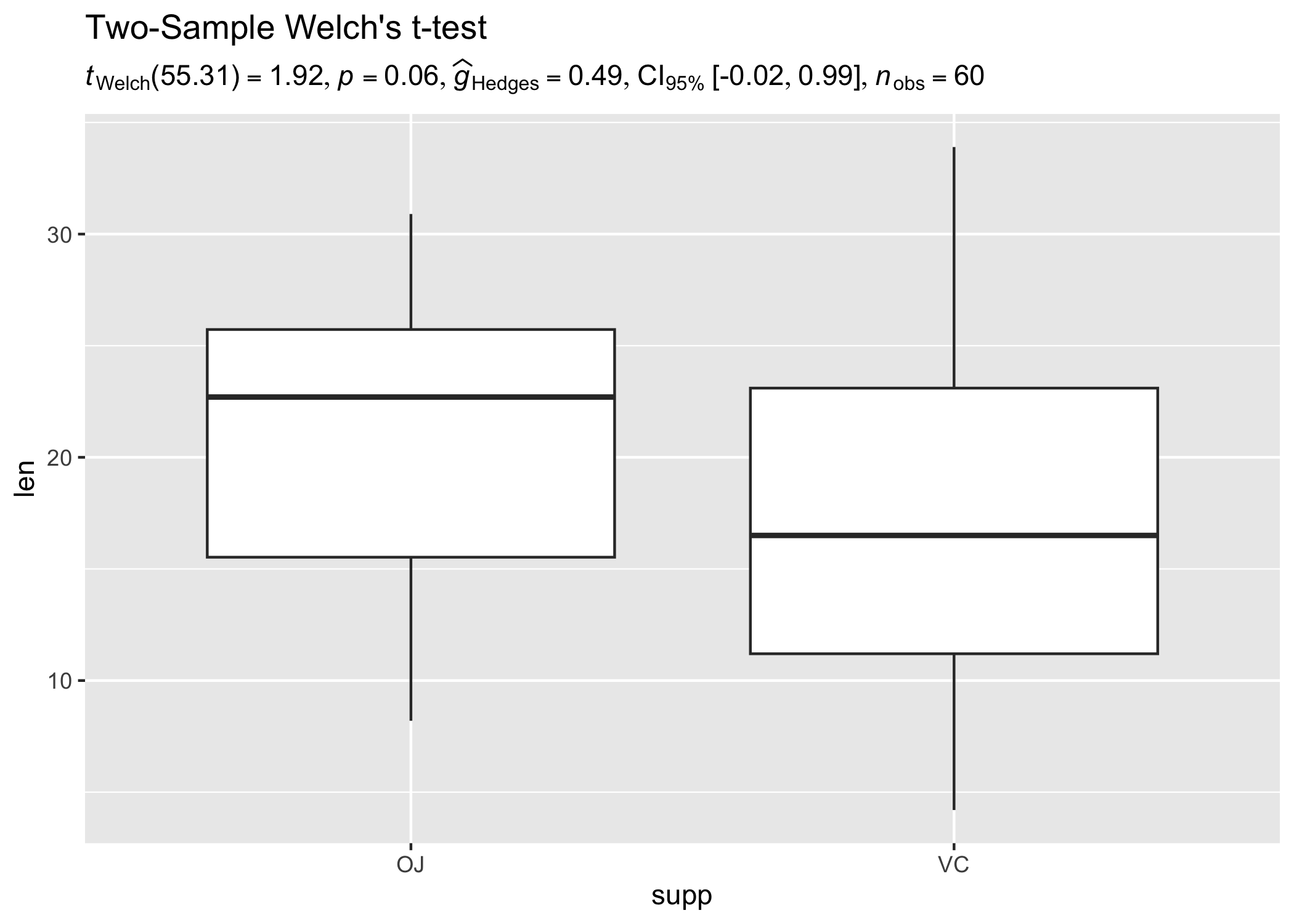
ggplot(ToothGrowth, aes(supp, len)) +
geom_half_boxplot() +
geom_beeswarm(beeswarmArgs = list(side = 1)) +
theme_ipsum_rc() +
# adding a subtitle with
labs(
title = "Two-Sample Welch's t-test",
subtitle = two_sample_test(ToothGrowth, supp, len)$expression[[1]]
)
Example with ggpubr:
# setup
set.seed(123)
library(ggplot2)
library(ggpubr)
library(statsExpressions)
# basic plot
gghistogram(
data.frame(
sex = factor(rep(c("F", "M"), each = 200)),
weight = c(rnorm(200, 55), rnorm(200, 58))
),
x = "weight",
add = "mean",
rug = TRUE,
fill = "sex",
palette = c("#00AFBB", "#E7B800"),
add_density = TRUE
) + # displaying stats results
labs(
title = "Yuen's two-sample test for trimmed means",
subtitle = two_sample_test(
data = data.frame(
sex = factor(rep(c("F", "M"), each = 200)),
weight = c(rnorm(200, 55), rnorm(200, 58))
),
x = sex,
y = weight,
type = "robust"
)$expression[[1]]
)

Another example with ggiraphExtra:
# setup
set.seed(123)
library(ggplot2)
library(ggiraphExtra)
library(gcookbook)
library(statsExpressions)
# plot
ggDot(heightweight, aes(sex, heightIn, fill = sex),
boxfill = "white",
binwidth = 0.4
) +
labs(
title = "Mann-Whitney test",
subtitle = two_sample_test(heightweight, sex, heightIn, type = "nonparametric")$expression[[1]]
)

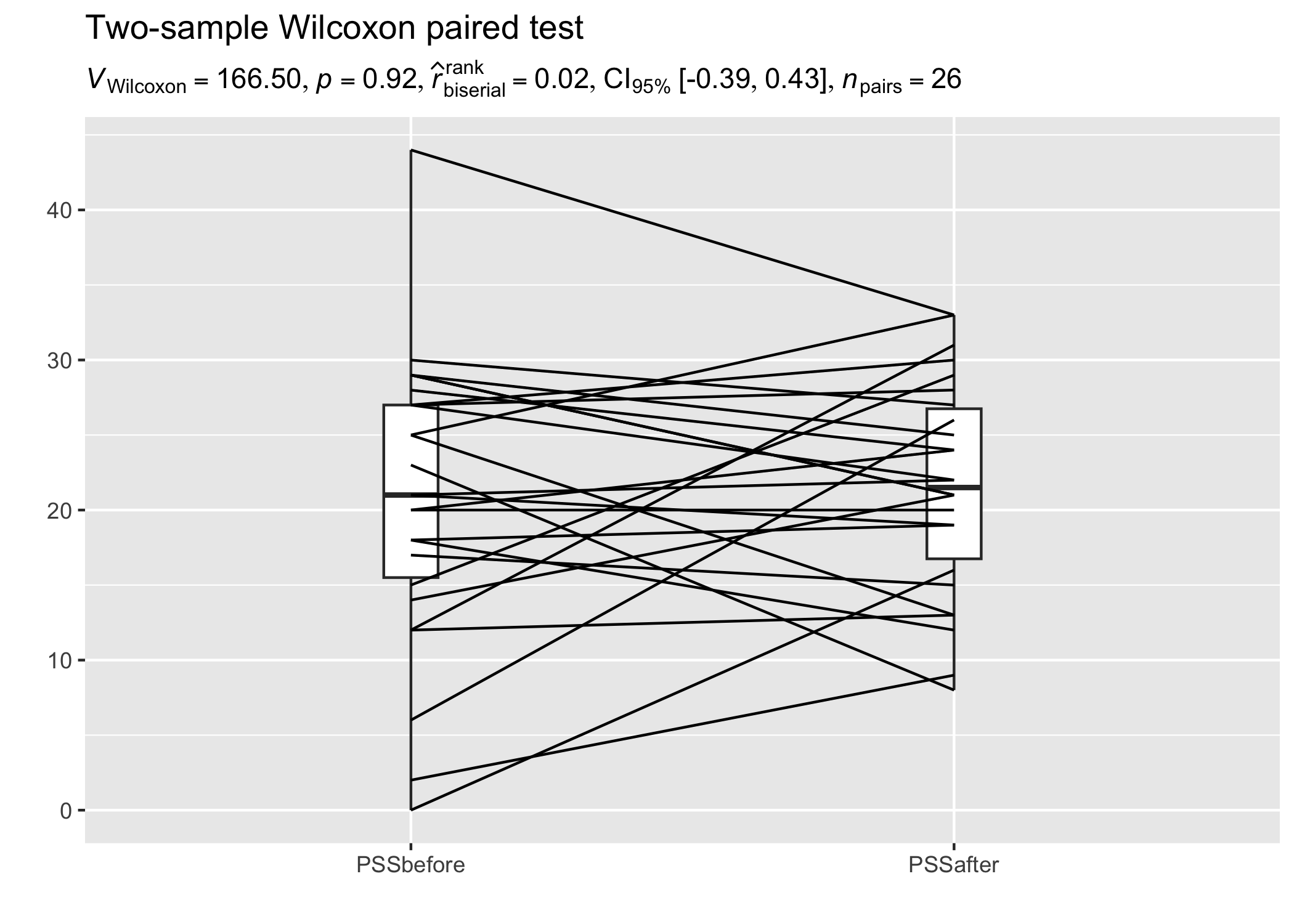
Within-subjects design
We can also have a look at a repeated measures design and the related expressions.
# setup
set.seed(123)
library(ggplot2)
library(statsExpressions)
library(tidyr)
library(PairedData)
data(PrisonStress)
# plot
paired.plotProfiles(PrisonStress, "PSSbefore", "PSSafter", subjects = "Subject") +
# `statsExpressions` needs data in the tidy format
labs(
title = "Two-sample Wilcoxon paired test",
subtitle = two_sample_test(
data = pivot_longer(PrisonStress, starts_with("PSS"), "PSS", values_to = "stress"),
x = PSS,
y = stress,
paired = TRUE,
subject.id = Subject,
type = "np"
)$expression[[1]]
)
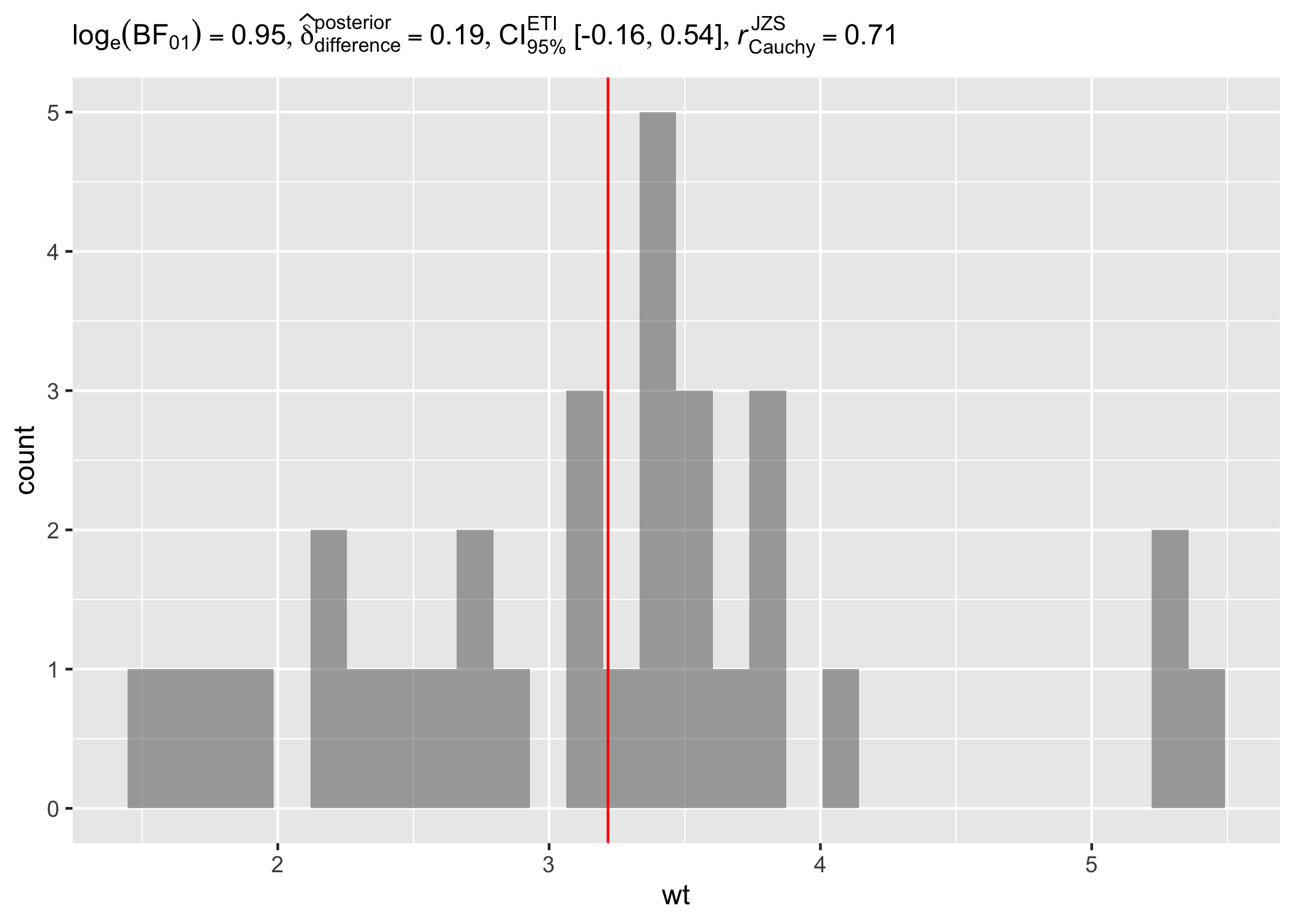
Example: Expressions for one-sample tests
# setup
set.seed(123)
library(ggplot2)
library(statsExpressions)
# creating a histogram plot
ggplot(mtcars, aes(wt)) +
geom_histogram(alpha = 0.5) +
geom_vline(xintercept = mean(mtcars$wt), color = "red") +
# adding a caption with a non-parametric one-sample test
labs(
title = "One-Sample Wilcoxon Signed Rank Test",
subtitle = one_sample_test(mtcars, wt, test.value = 3, type = "nonparametric")$expression[[1]]
)
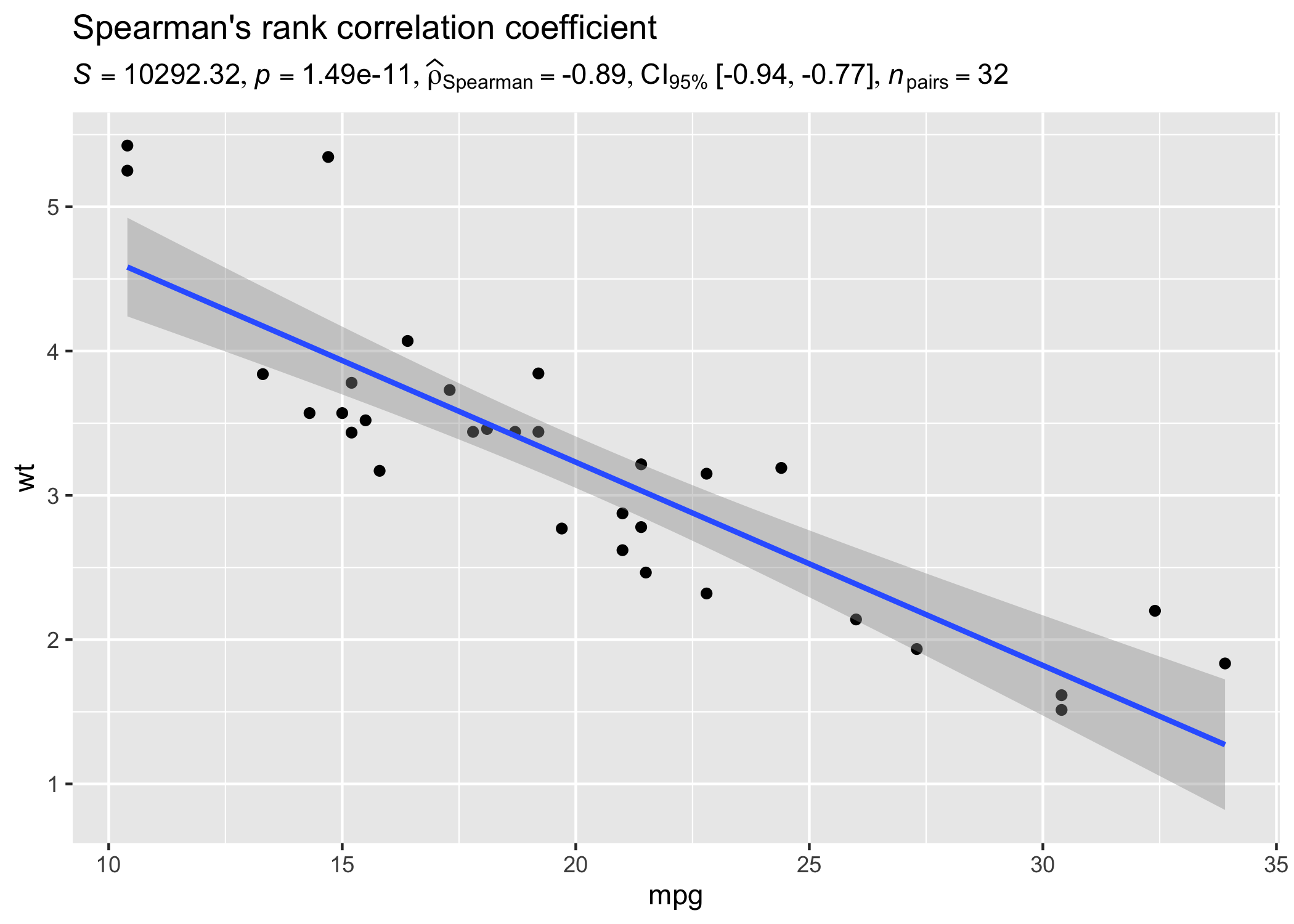
Example: Expressions for correlation analyses
Let’s look at another example where we want to run correlation analysis:
# setup
set.seed(123)
library(ggplot2)
library(statsExpressions)
# create a scatter plot
ggplot(mtcars, aes(mpg, wt)) +
geom_point() +
geom_smooth(method = "lm", formula = y ~ x) +
labs(
title = "Spearman's rank correlation coefficient",
subtitle = corr_test(mtcars, mpg, wt, type = "nonparametric")$expression[[1]]
)
Another example
# setup
set.seed(123)
library(ggplot2)
library(ggExtra)
library(statsExpressions)
# basic plot
p <-
ggplot(mtcars, aes(mpg, wt)) +
geom_point() +
geom_smooth(method = "lm") +
labs(
title = "Pearson's correlation coefficient",
subtitle = corr_test(mtcars, mpg, wt, type = "parametric")$expression[[1]]
)
# add
ggMarginal(p, type = "histogram", xparams = list(binwidth = 1, fill = "orange"))

Example: Expressions for contingency table analysis
For categorical/nominal data - one-sample:
# setup
set.seed(123)
library(ggplot2)
library(statsExpressions)
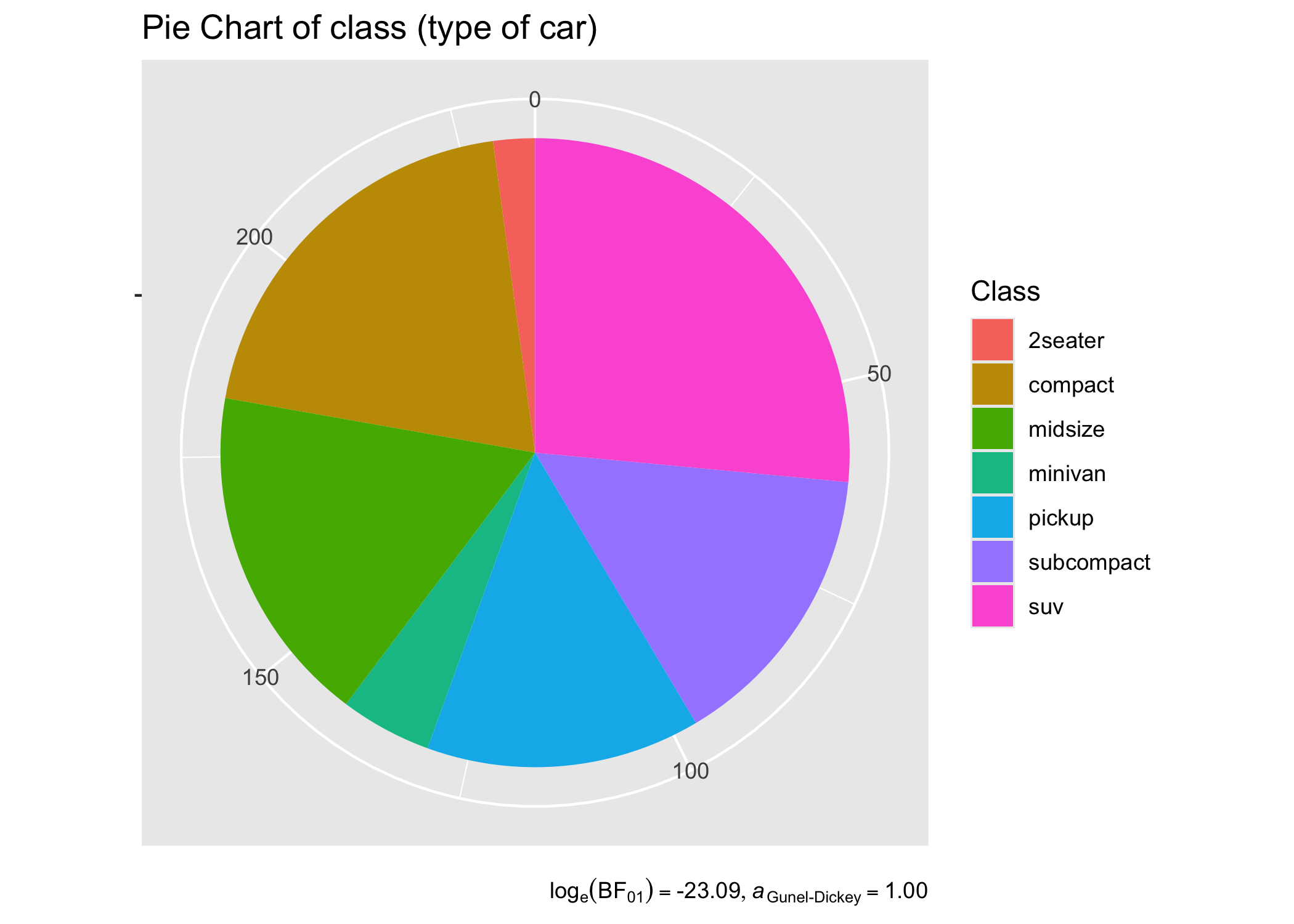
# basic pie chart
ggplot(as.data.frame(table(mpg$class)), aes(x = "", y = Freq, fill = factor(Var1))) +
geom_bar(width = 1, stat = "identity") +
theme(axis.line = element_blank()) +
# cleaning up the chart and adding results from one-sample proportion test
coord_polar(theta = "y", start = 0) +
labs(
fill = "Class",
x = NULL,
y = NULL,
title = "Pie Chart of class (type of car)",
subtitle = contingency_table(as.data.frame(table(mpg$class)), Var1, counts = Freq)$expression[[1]],
caption = "One-sample goodness of fit proportion test"
)
Another example of contingency tabs analysis:
# setup
set.seed(123)
library(moonBook)
library(ggiraphExtra)
library(statsExpressions)
# plot
ggSpine(
data = acs,
aes(x = Dx, fill = smoking),
addlabel = TRUE,
interactive = FALSE
) +
labs(
x = "diagnosis",
title = "Pearson's chi-squared contingency table test for counts",
subtitle = contingency_table(acs, Dx, smoking, paired = FALSE)$expression[[1]]
)

You can also use these function to get the expression in return without having to display them in plots:
# setup
set.seed(123)
library(ggplot2)
library(statsExpressions)
# Pearson's chi-squared test of independence
contingency_table(mtcars, am, cyl)$expression[[1]]
#> paste(chi["Pearson"]^2, "(", "2", ") = ", "8.74", ", ", italic("p"),
#> " = ", "0.013", ", ", widehat(italic("V"))["Cramer"], " = ",
#> "0.46", ", CI"["95%"], " [", "0.00", ", ", "0.78", "]", ", ",
#> italic("n")["obs"], " = ", "32")
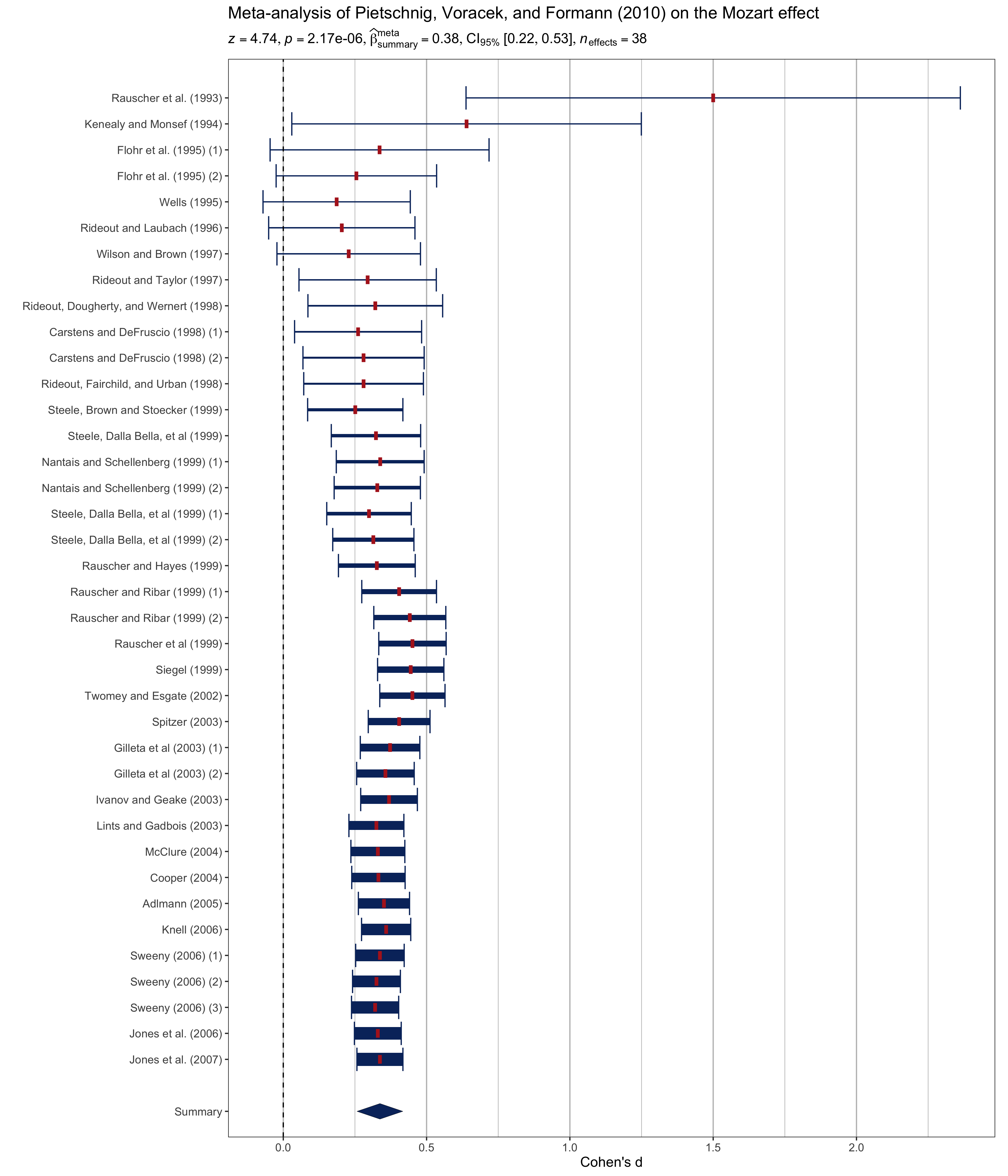
Example: Expressions for meta-analysis
# setup
set.seed(123)
library(metaviz)
library(ggplot2)
library(metaplus)
# meta-analysis forest plot with results random-effects meta-analysis
viz_forest(
x = mozart[, c("d", "se")],
study_labels = mozart[, "study_name"],
xlab = "Cohen's d",
variant = "thick",
type = "cumulative"
) + # use `statsExpressions` to create expression containing results
labs(
title = "Meta-analysis of Pietschnig, Voracek, and Formann (2010) on the Mozart effect",
subtitle = meta_analysis(dplyr::rename(mozart, estimate = d, std.error = se))$expression[[1]]
) +
theme(text = element_text(size = 12))
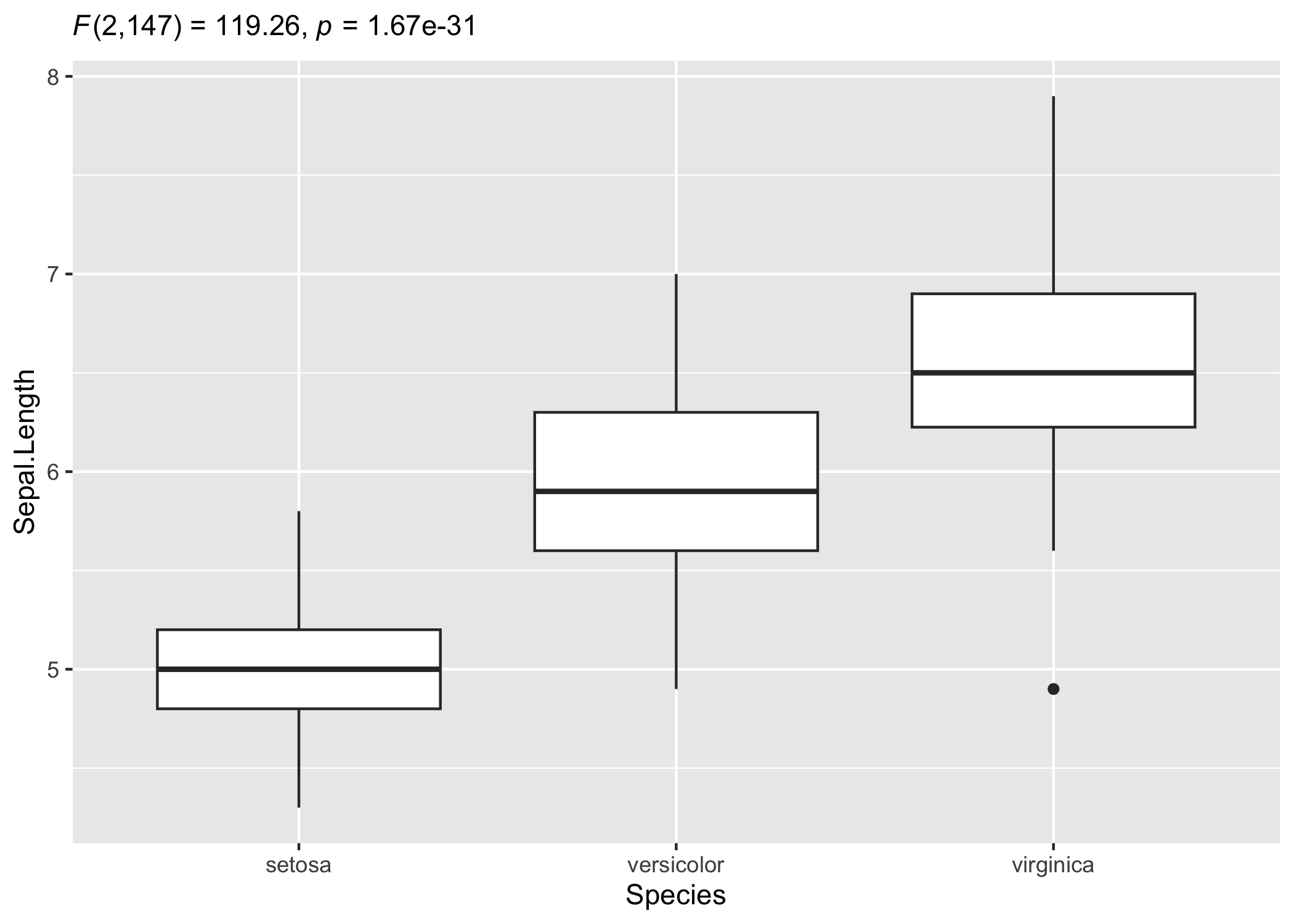
Customizing details to your liking
Sometimes you may not wish include so many details in the subtitle. In that case, you can extract the expression and copy-paste only the part you wish to include. For example, here only statistic and p-values are included:
# setup
set.seed(123)
library(ggplot2)
library(statsExpressions)
# extracting detailed expression
(res_expr <- oneway_anova(iris, Species, Sepal.Length, var.equal = TRUE)$expression[[1]])
#> paste(italic("F")["Fisher"], "(", "2", ",", "147", ") = ", "119.26",
#> ", ", italic("p"), " = ", "1.67e-31", ", ", widehat(omega["p"]^2),
#> " = ", "0.61", ", CI"["95%"], " [", "0.52", ", ", "0.68",
#> "]", ", ", italic("n")["obs"], " = ", "150")
# adapting the details to your liking
ggplot(iris, aes(x = Species, y = Sepal.Length)) +
geom_boxplot() +
labs(subtitle = ggplot2::expr(paste(
NULL, italic("F"), "(", "2",
",", "147", ") = ", "119.26", ", ",
italic("p"), " = ", "1.67e-31"
)))
Usage in ggstatsplot
Note that these functions were initially written to display results from
statistical tests on ready-made ggplot2 plots implemented in
ggstatsplot.
For detailed documentation, see the package website: https://indrajeetpatil.github.io/ggstatsplot/
Here is an example from ggstatsplot of what the plots look like when
the expressions are displayed in the subtitle-

Acknowledgments
The hexsticker was generously designed by Sarah Otterstetter (Max Planck Institute for Human Development, Berlin).
Code coverage
As the code stands right now, here is the code coverage for all primary functions involved: https://codecov.io/gh/IndrajeetPatil/statsExpressions/tree/master/R
Contributing
I’m happy to receive bug reports, suggestions, questions, and (most of
all) contributions to fix problems and add features. I personally prefer
using the GitHub issues system over trying to reach out to me in other
ways (personal e-mail, Twitter, etc.). Pull Requests for contributions
are encouraged.
Here are some simple ways in which you can contribute (in the increasing order of commitment):
-
Read and correct any inconsistencies in the documentation
-
Raise issues about bugs or wanted features
-
Review code
-
Add new functionality (in the form of new plotting functions or helpers for preparing subtitles)
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
