txie-93 / Gdynet
Labels
Projects that are alternatives of or similar to Gdynet
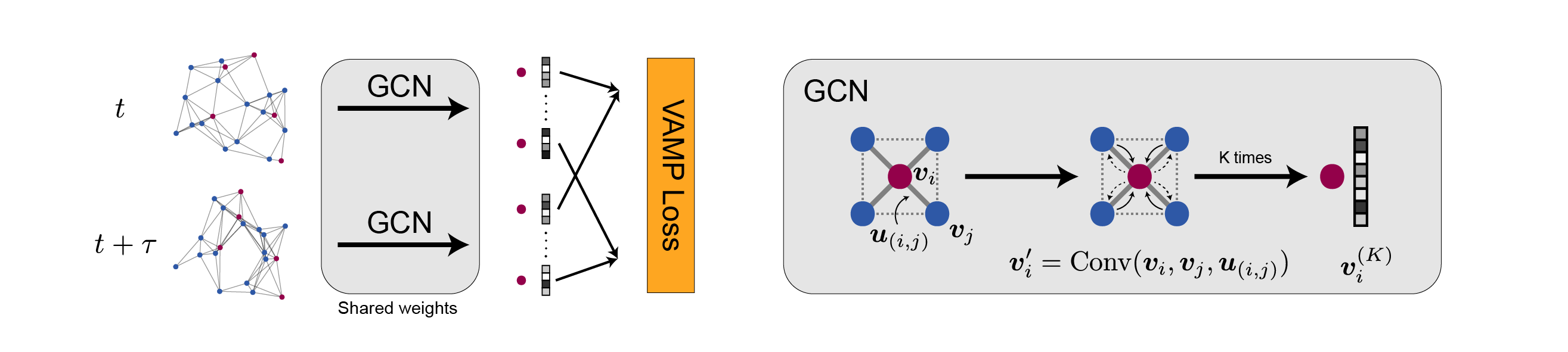
Graph Dynamical Networks
This software package implements the Graph Dynamical Networks (GDyNet) that automatically learns atomic scale dynamics for materials from molecular dynamics trajectories.
The following paper describes the details of the the method.
Graph Dynamical Networks for Unsupervised Learning of Atomic Scale Dynamics in Materials
Table of Contents
How to cite
Please cite the following work if you want to use GDyNet.
@article{xie_graph_2019,
title = {Graph dynamical networks for unsupervised learning of atomic scale dynamics in materials},
volume = {10},
issn = {2041-1723},
url = {http://www.nature.com/articles/s41467-019-10663-6},
doi = {10.1038/s41467-019-10663-6},
language = {en},
number = {1},
urldate = {2019-06-19},
journal = {Nature Communications},
author = {Xie, Tian and France-Lanord, Arthur and Wang, Yanming and Shao-Horn, Yang and Grossman, Jeffrey C.},
month = dec,
year = {2019},
pages = {2667}
}
Prerequisites
This package requires:
If you are new to Python, the easiest way of installing the prerequisites is via conda. After installing conda, run the following command to create a new environment named gdynet and install all prerequisites:
conda upgrade conda
conda create -n gdynet python=3.6 tensorflow pymatgen tqdm -c anaconda -c conda-forge
This creates a conda environment for running GDyNet. Before using GDyNet, activate the environment by:
source activate gdynet
Then, in directory gdynet, you can test if all the prerequisites are installed properly by running:
python main.py -h
python preprocess.py -h
This should display the help messages for main.py and preprocess.py. If you find no error messages, it means that the prerequisites are installed properly.
After you finished using GDyNet, exit the environment by:
source deactivate
Usage
Preprocess MD data
The GdyNets use dynamically constructed graphs to represent the atomic structures of materials in the MD trajectories. Unfortunately, we can't construct the graphs during the neural network training process because the graph construction algorithm is slow. So, we provide a preprocess.py to construct graphs from the MD trajectories as a preprocessing step.
First, you need to prepare your MD trajectories into a traj.npz file using numpy.savez_compressed. It has a dictionary-like data structure which can be queried for its list of arrays with fast IO. For a MD trajectory containing N atoms and F frames, the traj.npz file contains,
-
traj_coords:np.floatarrays with shape(F, N, 3), stores the cartesian coordinates of each atom in each frame. -
lattices:np.floatarrays with shape(F, 3, 3), stores the lattice matrix of the simulation box in each frame. In the lattice matrix, each row represents a lattice vector. -
atom_types:np.intarrays with shape(N,), stores the atomic number of each atom in the MD simulation. -
target_index:np.intarrays with shape(n,), stores the indexes (0-based) of the target atoms (n <= N). Note that here we assume the target atoms are just single atoms (like Li-ions) and each index repesents one target atom. We will support cases where target atoms are molecules (like water molecules) in the futhre.
Then, you can use the preprocess.py to preprocess the traj.npz. It will create a graph.npz file that contains the dynamically constructed graphs.
python preprocess.py traj.npz graph.npz
Note that the graph construction is slow especially for large MD trajectories. There two different graph construction algorithms implemented. The default --backend kdtree has a linear scaling but only works for orthogonal simulation box. For non-orthogonal simulation, use flag --backend direct which has a quadratic scaling. You can also take advantage of the multiprocessing with flag --n-workers. For other flags, checkout the help information with python preprocess.py -h.
Training the model
Before training the model, you should split your MD trajectory into three parts for training, validation, and testing. Then, following the steps in preprocess MD data, you can construct graphs for these three trajectories and obtain train-graph.npz, val-graph.npz, and test-graph.npz.
Now, you can start to train a GDyNet model. To keep everything clean, we create a new folder training to store all the intermediate files.
mkdir training
Then, you can use the main.py to train your model.
python main.py --train-flist train-graph.npz --val-flist val-graph.npz --test-flist test-graph.npz --job-dir training/
There are several common issues that may prevent the training from success.
- If the loss becomes
NaN, it usually means the learning rate is too high. Try decrease it with flag--lr. - If there is GPU memory error, you may want to decrease the batch size with flag
-b. - Make sure that the
--modematches the graph construction algorithm. - You can also try different
--n-classes,--tau,--atom-fea-len, and--n-convto get the best performance. For other flags, usepython main.py -hfor helps.
Visualize the results
After the training, you can visualize the dynamical model using the functions in the postprocess module. We provide a jupyter notebook to demonstrate how to visualize the results for the toy system from the paper.
Data
To reproduce our results or use the data from our paper, see these instructions.
Authors
This software was primarily written by Tian Xie who was advised by Prof. Jeffrey Grossman.
The VAMP loss module is modified on top of the VAMPnet project by A. Mardt, et al. The PeriodicKDTree module is modifed on top of the periodic_kdtree project by P. Varilly.
License
Graph Dynamical Networks is released under the MIT license.