napari-aicsimageio
AICSImageIO bindings for napari
Features
- Supports reading metadata and imaging data for:
OME-TIFFTIFFCZI(Zeiss)LIF(Leica)ND2(Nikon)DV(DeltaVision)- Any formats supported by aicsimageio
- Any formats supported by bioformats
SLD(Slidebook)SVS(Aperio)- Full List
- Any additional format supported by imageio
PNGJPGGIFAVI- Full List
While upstream aicsimageio is released under BSD-3 license, this plugin is released under GPLv3 license because it installs all format reader dependencies.
Installation
Stable Release: pip install napari-aicsimageio or conda install napari-aicsimageio -c conda-forge
Development Head: pip install git+https://github.com/AllenCellModeling/napari-aicsimageio.git
Plugin Variants
There are two variants of this plugin that are added during installation:
aicsimageio-in-memory, which reads an image fully into memoryaicsimageio-out-of-memory, which delays reading ZYX chunks until required. This allows for incredibly large files to be read and displayed.
Examples of Features
General Image Reading
All image file formats supported by aicsimageio will be read and all raw data will be available in the napari viewer.
In addition, when reading an OME-TIFF, you can view all OME metadata directly in the
napari viewer thanks to ome-types.
Multi-Scene Selection
When reading a multi-scene file, a widget will be added to the napari viewer to manage scene selection (clearing the viewer each time you change scene or adding the scene content to the viewer) and a list of all scenes in the file.
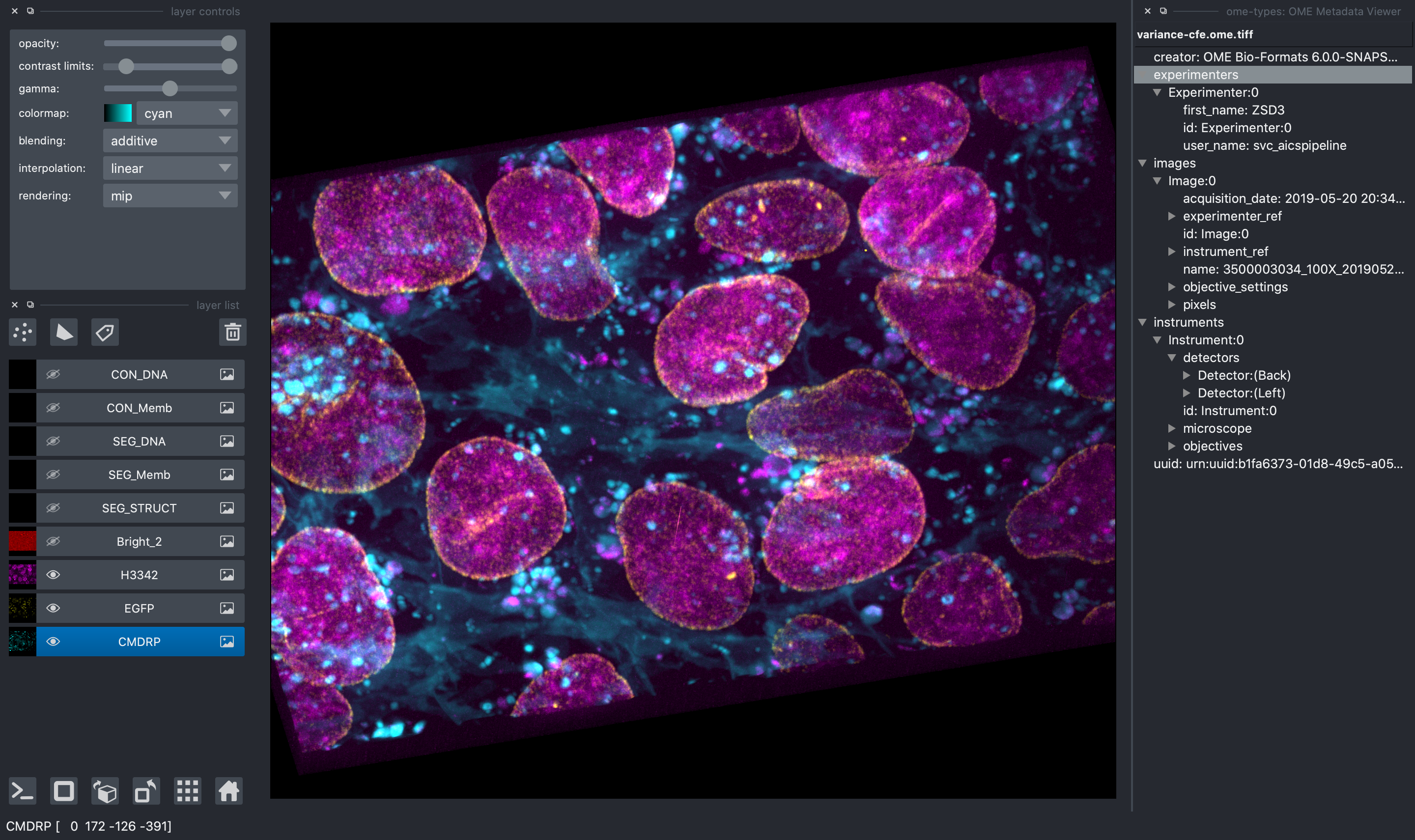
Access to the AICSImage Object and Metadata
You can access the AICSImage object used to load the image pixel data and
image metadata using the built-in napari console:
img = viewer.layers[0].metadata["aicsimage"]
img.dims.order # TCZYX
img.channel_names # ["Bright", "Struct", "Nuc", "Memb"]
img.get_image_dask_data("ZYX") # dask.array.ArrayThe napari layer metadata dictionary also stores a shorthand for the raw image metadata:
viewer.layers[0].metadata["raw_image_metadata"]The metadata is returned in whichever format is used by the underlying
file format reader, i.e. for CZI the raw metadata is returned as
an xml.etree.ElementTree.Element, for OME-TIFF the raw metadata is returned
as an OME object from ome-types.
Lastly, if the underlying file format reader has an OME metadata conversion function,
you may additionally see a key in the napari layer metadata dictionary
called "ome_types". For example, because the AICSImageIO
CZIReader and BioformatsReader both support converting raw image metadata
to OME metadata, you will see an "ome_types" key that stores the metadata transformed
into the OME metadata model.
viewer.layers[0].metadata["ome_types"] # OME object from ome-typesMosaic Reading
When reading CZI or LIF images, if the image is a mosaic tiled image, napari-aicsimageio
will return the reconstructed image:
Development
See CONTRIBUTING.md for information related to developing the code.
For additional file format support, contributed directly to
AICSImageIO.
New file format support will become directly available in this
plugin on new aicsimageio releases.
Citation
If you find aicsimageio (or napari-aicsimageio) useful, please cite as:
AICSImageIO Contributors (2021). AICSImageIO: Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Pure Python [Computer software]. GitHub. https://github.com/AllenCellModeling/aicsimageio
Free software: GPLv3



![napari viewer with console open showing viewer.layers[0].metadata](https://raw.githubusercontent.com/AllenCellModeling/napari-aicsimageio/main/images/console-access.png)