alistairewj / Sepsis3 Mimic
Labels
Projects that are alternatives of or similar to Sepsis3 Mimic
Sepsis-3 in MIMIC-III 
This is the code repository associated with A Comparative Analysis of Sepsis Identification Methods in an Electronic Database
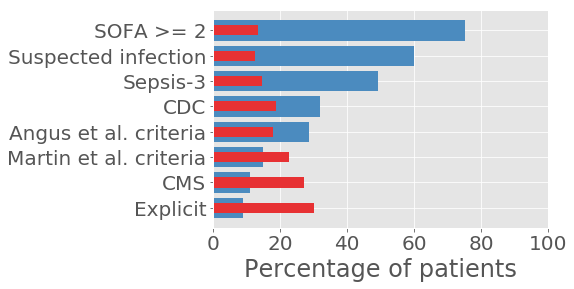
The publication assessed five methods of identifying sepsis in electronic health records, and found that all five had varying cohort sizes and severity of illness as measured by in-hospital mortality rate. The results are best summarized by the following figure:
Above, we can see that, as we change the criteria used to define sepsis, the percentage of patients who satisfy the criteria decreases (blue bars) and the percent mortality of that cohort increases (red bars). For more detail please see the paper.
Acknowledgement
If you find the code useful, we would appreciate acknowledging our work with a citation: either the code directly (using the DOI badge from Zenodo), the paper summarizing the work, or both!
@article{johnson2018comparative,
title={A Comparative Analysis of Sepsis Identification Methods in an Electronic Database},
author={Johnson, Alistair EW and Aboab, Jerome and Raffa, Jesse D and Pollard, Tom J and Deliberato, Rodrigo O and Celi, Leo A and Stone, David J},
journal={Critical care medicine},
volume={46},
number={4},
pages={494--499},
year={2018},
publisher={Wolters Kluwer}
}
@misc{alistair_johnson_2018_1256723,
author = {Alistair Johnson and Tom Pollard},
title = {sepsis3-mimic},
version = {1.0.0},
publisher = {Zenodo},
month = may,
year = 2018,
doi = {10.5281/zenodo.1256723},
url = {https://doi.org/10.5281/zenodo.1256723}
}
Reproducing the results of the above study
Reproducing the study can be done as follows:
- Installing necessary Python dependencies
- Generate or acquire the CSV files which are used for analysis by...
- Running the
sepsis-3-get-data.ipynbnotebook from start to finish - ... or downloading the CSV files from the MIMIC-III Derived Data Repository and analyzing those files
- Running the analysis in
sepsis-3-main.ipynb
The following instructions have been tested on Ubuntu 16.04, but should be relatively straightforward to adapt to other systems.
1. Clone the repository and install necessary Python dependencies
You will need a local copy of the code in this repository. The easiest way to acquire this is to use git to clone the data locally. If using Ubuntu, you can install git easily with:
sudo apt-get install git
Next, clone the repository with the --recursive flag as it relies on a distinct repository (mimic-code):
git clone https://github.com/alistairewj/sepsis3-mimic sepsis3-mimic --recursive
If you already have the repository cloned on your local computer, but you didn't use the --recursive flag, you can clone the submodule easily:
cd sepsis3-mimic
git submodule update --init --recursive
(Optional, recommended): Create a virtual environment for this repository. I like to use virtualenvwrapper as it makes managing virtual environments easier. After installing virtualenvwrapper, run: mkvirtualenv --python=python3 sepsis3-py3
If you are running jupyter notebook in a different environment (e.g. you have a system level install of jupyter currently running, but want to use this virtual environment), then you can add this virtual environment as a kernel specification. From outside your virtual environment, run: python -m ipykernel install --user --name sepsis3-py3 --display-name "sepsis3-py3". You can read more about this here. After installing the kernel specification, you may need to reload any active notebooks in order to see the kernel as an option.
Finally, using a package manager for Python (pip), you can run the following from the root directory of this repository to install all necessary python packages:
pip install -r requirements.txt
2. Acquire CSVs from a database with MIMIC-III
There are two options: (a) regenerate the CSVs from the original MIMIC-III database, or (b) download pre-generated CSVs from PhysioNetWorks. Both will require approval to access MIMIC-III - you can also read more about getting access to MIMIC-III.
(a) Regenerate the CSVs from a PostgreSQL database with MIMIC-III
The sepsis-3-get-data.ipynb notebook runs through the process of exporting the data from the database and writing it to CSV files. This notebook requires:
- PostgreSQL version 9.4 or later
- The MIMIC-III database installed in PostgreSQL
If you do not have the above, you can follow the instructions on this page to access and install MIMIC-III.
Once you have the database setup, you will need to generate the CSV files. The easiest way is to run through the sepsis-3-get-data.ipynb notebook: this will call the query/make-tables.sql script and generate the necessary tables on the database. Alternatively, you can run this script directly, and the notebook will recognize that the final sepsis3 table already exists. If running directly using psql, you can call:
cd query
psql
set search_path to public,mimiciii;
\i make-tables.sql
Either way, the generation of all the tables can take anywhere from 10 minutes to about an hour, depending on your system. You may see a lot of NOTICE warnings: don't worry about them. The query logic is "check if the table exists, and if it does, drop it". These warnings indicate that the table did not exist (and nor would you expect it to on a fresh install!).
(b) Download the CSVs from the MIMIC-III Derived Data repository
The data files can be downloaded from the MIMIC-III Derived Data Repository.
If you have a PhysioNetWorks account which has been approved for access to MIMIC-III, access the sepsis3-mimic derived data repository here, supplying your username and password when prompted.
The data file is "sepsis3-data.zip". By default, the code expects the data to be extracted in the data subfolder of this repository.
3. Run analysis
sepsis-3-main.ipynb - this analyzes the data and reports all results found in the paper. It assumes the data is available in the data subfolder of this directory - all notebooks do this, and you can change it if you like by modifying the data_path variable at the top of each script.
(Optional) Supplemental Material
Results presented in the supplemental material can be regenerated using the supplemental-material.ipynb file.
(Optional) Other notebooks
There are a number of other notebooks: venn-diagrams.ipynb unsurprisingly generates many Venn diagrams, criteria-over-time.ipynb and the appendix subfolder contains a number of notebooks/R scripts which contain some interesting analyses but may not work out of the box.
Pull requests welcome! :)