README
The strcode (short for structuring code) package contains tools to organize and abstract your code better. It consists of
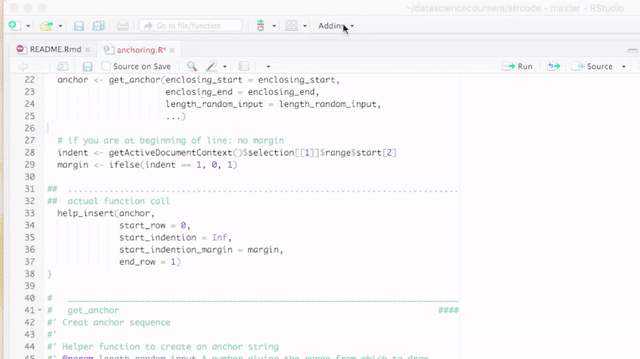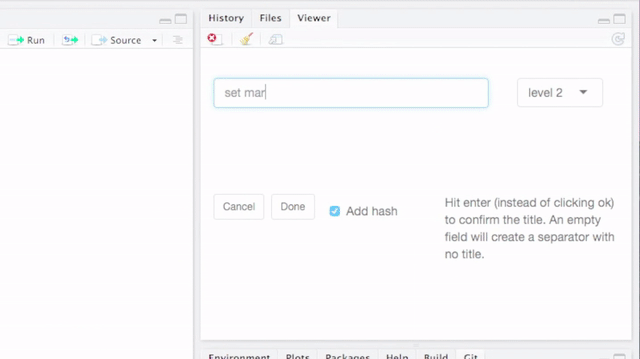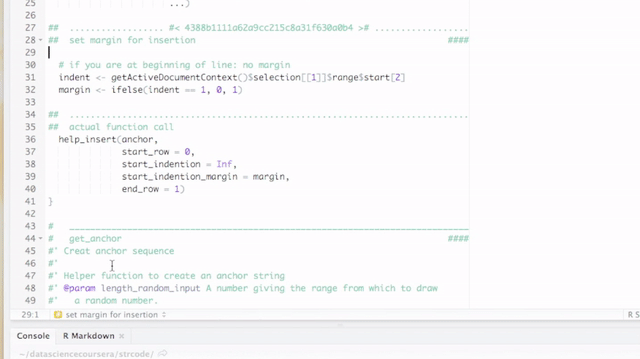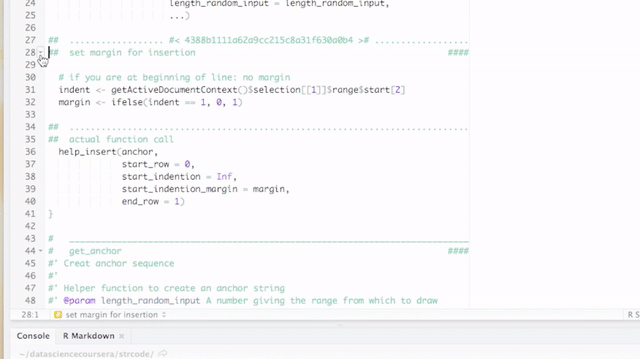
- An RStudio Add-in that lets you quickly add code block separators and titles (possibly with unique identifiers) to divide your work into sections. The titles are recognized as sections by RStudio, which enhances the coding experience further.
- A function
sum_strthat summarizes the code structure based on the separators and their comments added with the Add-in. For one or more files, it can cat the structure to the console or a file. - An RStudio Add-in that lets you insert a code anchor, that is, a hash sequence which can be used to uniquely identify a line in a large code base.
Installation
You can install the package from GitHub.
# install.packages("devtools")
devtools::install_github("lorenzwalthert/strcode")Structuring code
We suggest three levels of granularity for code structuring, whereas higher-level blocks can contain lower-level blocks.
- level 1 sections, which are high-level blocks that can be separated as follows:
# ____________________________________________________________________________
# A title ####- level 2 sections, which are medium-level blocks that can be separated as follows:
## ............................................................................
## A subtitle ####- level 3 sections, which are low-level blocks that can be separated as follows:
### .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
### One more ####You can notice from the above that
- The number of
#used in front of the break character (___,...,. .) corresponds to the level of granularity that is separated. - The breaks characters
___,...,. .were chosen such that they reflect the level of granularity, namely___has a much higher visual density than. .. - Each block has an (optional) short title on what that block is about.
- Every title ends with
####. Therefore, the titles are recognized by RStudio as sections. This has the advantages that you can get a quick summary of your code in Rstudio's code pane and you can fold sections as you can fold code or function declarations or if statements. See the pictures below for details.
The separators all have length 80. The value is looked up in the global option strcode$char_length and can therefore be changed by the user.
By default, breaks and titles are inserted via a Shiny Gadget, but this default can be overridden by setting the option strcode$insert_with_shiny to FALSE and hence only inserting the break.
Anchoring sections
Sometimes it is required to refer to a code section, which can be done by title. A better way, however, is to use a unique hash sequence - let us call it a code anchor - to create an arguably unique reference to that section. A code anchor in strcode is enclosed by #< and ># so all anchors can be found using regular expressions. You can add section breaks that include a hash. That might look like this:
## .................. #< 685c967d4e78477623b861d533d0937a ># ..................
## An anchored section ####Insert a code anchor
Code anchors might prove helpful in other situations where one want to anchor a single line. That is also possible with strcode. An example of a code anchor is the following:
#< 56f5139874167f4f5635b42c37fd6594 >#
this_is_a_super_important_but_hard_to_describe_line_so_let_me_anchor_itThe hash sequences in strcode are produced with the R package digest.
Summarizing code
Once code has been structured by adding sections (as above), it can easily be summarized or represented in a compact and abstract form. This is particularly handy when the codebase is large, when a lot of people work on the code or when new people join a project. The function sum_str is designed for the purpose of extracting separators and respective comments, in order to provide high level code summaries. It is highly customizable and flexible, with a host of options. Thanks to RStudio's API, you can even create summaries of the file you are working on, simply by typing sum_str() in the console. The file presented in the example section below can be summarized as follows:
sum_str(path_in = "placeholder_code/example.R",
file_out = "",
width = 40,
granularity = 2,
lowest_sep = FALSE,
header = TRUE)
#> Summarized structure of placeholder_code/example.R
#>
#> line level section
#> 2 # _
#> 3 # function test
#> 6 ## -A: pre-processing
#> 57 ## B: actual function
#> 83 # ____________________________________
#> 84 # function test2
#> 87 ## A: pre-processing
#> 138 ## B: actual function
#> 169 ## testpath_inspecifies a directory or filenames for looking for content to summarize.file_outindicates where to dump the output.widthgives the width of the output in characters.granularity = 2indicates that we want two of three levels of granularity to be contained in the summary and don't include level 3 comments.- Similarly, we use
lowest_sep = FALSEto indicate that we want lowest separators (givengranularity) to be omitted between the titles of the sections. headerwas set toTRUE, so the column names were reported as well. Note that they are slightly off since knitr uses a different tab length. In the R console and more imporantly in the outputed file, they are aliged.
Example of improved legibility
To demonstrate the improvement in legibility, we give an extended example with some placeholder code.
# ____________________________________________________________________________
# function test ####
test <- function(x) {
## ............................................................................
## A: pre-processing ####
### .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
### a: assertive tests ####
# x
if(missing(x) || is.null(x)){
x <- character()
}
assert(
# use check within assert
check_character(x),
check_factor(x),
check_numeric(x)
)
# levels
if(!missing(levels)){
assert(
check_character(levels),
check_integer(levels),
check_numeric(levels))
levels <- na.omit(levels)
}
# labels
if(!missing(labels)){
assert(
check_character(labels),
check_numeric(labels),
check_factor(labels)
)
}
### .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
### b: coercion / remove missing ####
x <- as.character(x)
uniq_x <- unique(na.omit(x), nmax = nmax)
### .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
### c: warnings ####
if(length(breaks) == 1) {
if(breaks > max(x) - min(x) + 1) {
stop("range too small for the number of breaks specified")
}
if(length(x) <= breaks) {
warning("breaks is a scalar not smaller than the length of x")
}
}
## ............................................................................
## B: actual function ####
variable < -paste("T", period, "nog_", sector, sep = "")
variable <- paste(variable, "==", 1, sep = "")
arg<-substitute(variable)
r<-eval(arg, idlist.data[[1]])
a<<-1
was_factor <- FALSE
if (is.factor(yes)) {
yes <- as.character(yes)
was_factor <- TRUE
}
if (is.factor(no)) {
no <- as.character(no)
was_factor <- TRUE
}
out <- ifelse(test, yes, no)
if(was_factor) {
cfactor(out)
} else {
out
}
## ............................................................................
}
# ____________________________________________________________________________
# function test2 ####
test2 <- function(x) {
## ............................................................................
## A: pre-processing ####
### .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
### a: assertive tests ####
# x
if(missing(x) || is.null(x)){
x <- character()
}
assert(
# use check within assert
check_character(x),
check_factor(x),
check_numeric(x)
)
# levels
if(!missing(levels)){
assert(
check_character(levels),
check_integer(levels),
check_numeric(levels))
levels <- na.omit(levels)
}
# labels
if(!missing(labels)){
assert(
check_character(labels),
check_numeric(labels),
check_factor(labels)
)
}
### .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
### b: coercion / remove missing ####
x <- as.character(x)
uniq_x <- unique(na.omit(x), nmax = nmax)
### .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
### c: warnings ####
if(length(breaks) == 1) {
if(breaks > max(x) - min(x) + 1) {
stop("range too small for the number of breaks specified")
}
if(length(x) <= breaks) {
warning("breaks is a scalar not smaller than the length of x")
}
}
## ............................................................................
## B: actual function ####
variable < -paste("T", period, "nog_", sector, sep = "")
variable <- paste(variable, "==", 1, sep = "")
arg<-substitute(variable)
r<-eval(arg, idlist.data[[1]])
a<<-1
was_factor <- FALSE
if (is.factor(yes)) {
yes <- as.character(yes)
was_factor <- TRUE
}
if (is.factor(no)) {
no <- as.character(no)
was_factor <- TRUE
}
out <- ifelse(test, yes, no)
if(was_factor) {
cfactor(out)
} else {
out
}
## ............................................................................
}