RuiShu / Vae Clustering
Labels
Projects that are alternatives of or similar to Vae Clustering
VAE-Clustering
A collection of experiments that shines light on VAE (containing discrete latent variables) as a clustering algorithm.
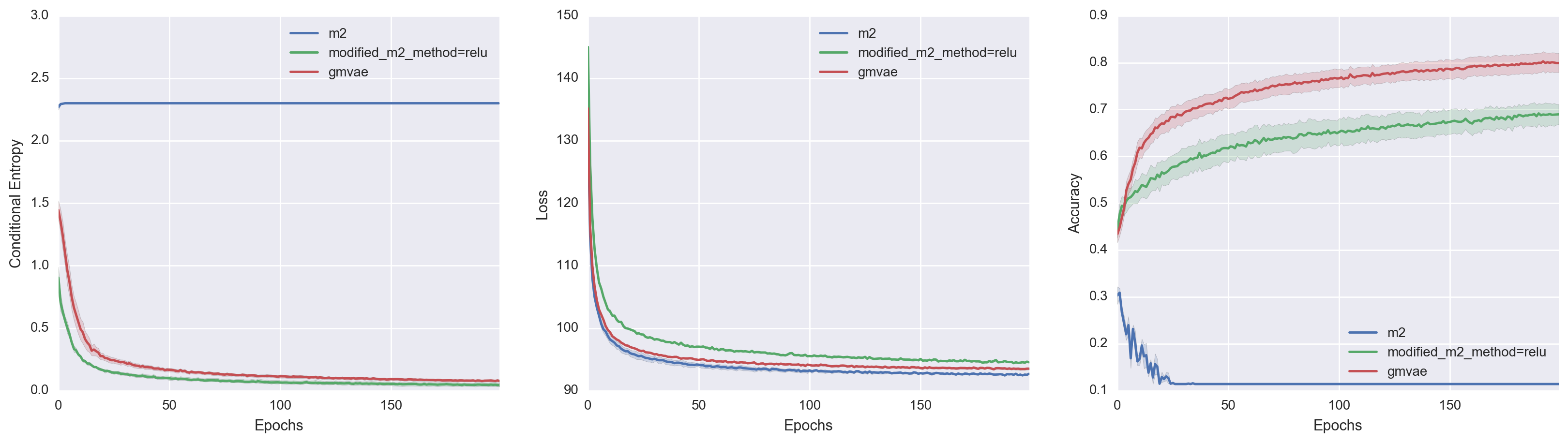
We evaluate the unsupervised clustering performance of three closely-related sets of deep generative models:
- Kingma's M2 model
- A modified-M2 model that implicitly contains a non-degenerate Gaussian mixture latent layer
- An explicit Gaussian Mixture VAE model
Details about the three models and why to compare them are provided in this blog post.
Results
M2 performs poorly as an unsupervised clustering algorithm. We suspect this is attributable to conflicting wishes to use the categorical variable as part of the generative model versus the inference model. By implicitly enforcing the a hidden layer to have a proper Gaussian mixture distribution, the modified-M2 model tips the scale in favor of using the categorical variable as part of the generative model. By using an explicit Gaussian Mixture VAE model, we achieve enable better inference, which leads to higher stability during training and even a stronger incentive to use the categorical variable in the generative model.
Code set-up
The experiments are implemented using TensorFlow. Since all of the three aforementioned models share very similar formulations, the shared subgraphs are placed in shared_subgraphs.py. The utils.py file contains some additional functions used during training. The remaining *.py files simply implement the three main model classes and other variants that we tried.
We recommend first reading the Jupyter Notebook on nbviewer in the Chrome browser.
Dependencies
- tensorflow
- tensorbayes
- numpy
- scipy