ElieAljalbout / Clustering With Deep Learning
Programming Languages
Projects that are alternatives of or similar to Clustering With Deep Learning
Deep Learning for Clustering
Code for project "Deep Learning for Clustering" under lab course "Deep Learning for Computer Vision and Biomedicine" - TUM. Depends on numpy, theano, lasagne, scikit-learn, matplotlib.
Contributors
- Mohd Yawar Nihal Siddiqui
- Elie Aljalbout
- Vladimir Golkov (Supervisor)
Related Papers:
This repository is an implementation of the paper : Elie Aljalbout, Vladimir Golkov, Yawar Siddiqui, Daniel Cremers "Clustering with Deep Learning: Taxonomy and new methods"
Usage
Use the main script for training, visualizing clusters and/or reporting clustering metrics
python main.py <options>
| Option | |
|---|---|
-d DATASET_NAME, --dataset DATASET_NAME |
(Required) Dataset on which autoencoder is to be trained trained, or metrics/visualizations are to be performed [MNIST,COIL20] |
-a ARCH_IDX, --architecture ARCH_IDX |
(Required) Index of architecture of autoencoder in the json file (archs/) |
--pretrain EPOCHS |
Pretrain the autoencoder for specified #epochs specified by architecture on specified dataset |
--cluster EPOCHS |
Refine the autoencoder for specified #epochs with clustering loss, assumes that pretraining results are available |
--metrics |
Report k-means clustering metrics on the clustered latent space, assumes pretrain and cluster based training have been performed |
--visualize |
Visualize the image space and latent space, assumes pre-training and cluster based training have been performed |
Project Structure
| Folder / File | Description |
|---|---|
| archs | Contains json files specifying architectures for autoencoder networks used. File mnist.json contains architectures for MNIST dataset. We use the second architecture for the reported results (command line argument -a 1) |
| coil, mnist | Contains the datasets COIL20 and MNIST respectively |
| logs | Output folder for logs generated by the scripts. Named by date and time of script execution |
| plots | Scatter plots showing the raw, pre-trained latent space, and the final latent space clusters |
| saved_params | Contains saved network parameters and saved representation of inputs in latent space |
| custom_layers.py | Custom lasagne layers, Unpool2D - which performs inverse max pooling by replicating input pixels as dictated by the filter size, and the ClusteringLayer - a layer that outputs soft cluster assignments based on k-means cluster distance |
| main.py | The main python script for training and evaluating the network |
| misc.py | Contains dataset handlers and other utility methods |
| network.py | Contains classes for parsing and building the network from json files and also for training the network |
Autoencoder Builder
We've implemented a NetworkBuilder class that can be used to quickly describe the architecture of an autoencoder through a json file. The json specification of the architecture is a dictionary with the following fields
| Field | Description |
|---|---|
| name | Name identifier given to the architecture, used for file naming while saving parameters |
| batch_size | Batch size to be used while training the network |
| use_batch_norm | Whether to use batch normalization for convolutional/deconvolutional layers |
| network_type | Type of network - convolutional or fully connected |
| layers | A list describing the encoder part of the autoencoder |
Further, each item in the layers list is a dictionary with the following fields
| Field | Description |
|---|---|
| type | Can be Input, Conv2D, MaxPool2D, MaxPool2D*, Dense, Reshape, Deconv2D |
| num_filters | For Conv2D/MaxPool2D/MaxPool2D*/Deconv2D layers this field specifies number of filters |
| filter_size | Dimensions of kernel for the above layers |
| num_units | For Dense layer number of hidden units |
| non_linearity | Non-Linearity function used at output of the layer |
| conv_mode | Can be used to specify the convolution mode like same, valid etc. for convolutional layers |
| output_non_linearity | If you want a different non linearity function at the output than the one which would be obtained by mirroring |
Only the encoder part of the autoencoder needs to be specified, the decoder will be automatically generated by the class.
Example of a network description
{
"name": "c-5-6_p_c-5-16_p_c-4-120",
"use_batch_norm": 1,
"batch_size": 100,
"layers": [
{
"type": "Input",
"output_shape":[1, 28, 28]
},
{
"type": "Conv2D",
"num_filters": 50,
"filter_size": [5, 5],
"non_linearity": "rectify"
},
{
"type": "MaxPool2D*",
"filter_size": [2, 2]
},
{
"type": "Conv2D",
"num_filters": 50,
"filter_size": [5, 5],
"non_linearity": "rectify"
},
{
"type": "MaxPool2D*",
"filter_size": [2, 2]
},
{
"type": "Conv2D",
"num_filters": 120,
"filter_size": [4, 4],
"non_linearity": "linear"
}
]
}
This would generate the network
50[5x5] 50[5x5]_bn max[2x2] 50[5x5] 50[5x5]_bn max[2x2] **120[4x4] 120[4x4]_bn **50[4x4] 50[4x4]_bn ups*[2x2] 50[5x5] 50[5x5]_bn ups*[2x2] 1[5x5]
Experiments and Results
We trained and tested the network on two datasets - MNIST and COIL20
| Dataset | Image size | Number of samples | Number of clusters |
|---|---|---|---|
| MNIST | 28x28x1 | 60000 | 10 |
| COIL20 | 128x128x1 | 1440 | 20 |
Clustering was performed with two different loss functions -
- Loss =
KL-Divergence(soft assignment distribution, target distribution) + Autoencoder Reconstruction loss, where the target distribution is a distribution that improves cluster purity and puts more emphasis on data points assigned with a high confidence. For more details check out the DEC paper [1]. - Loss =
k-Means loss + Autoencoder Reconstruction loss
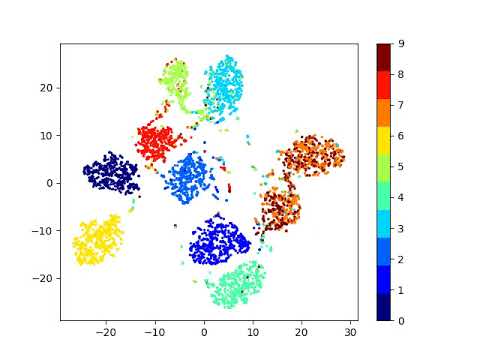
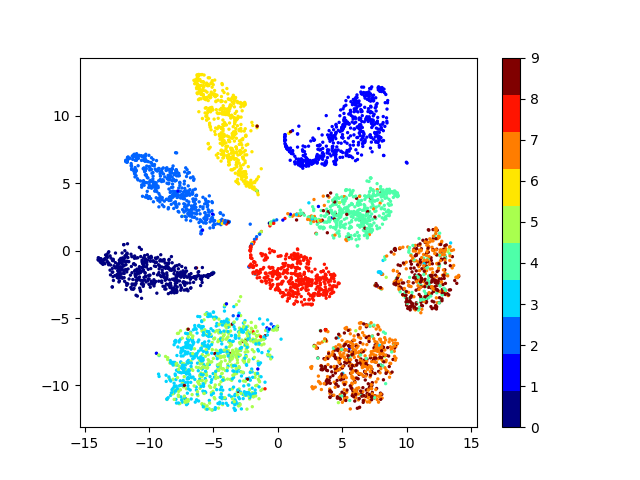
MNIST
Our network
| Clustering space | Clustering Accuracy | Normalized Mutual Information |
|---|---|---|
| Image pixels | 0.542 | 0.480 |
| Autoencoder | 0.760 | 0.667 |
| Autoencoder + k-Means Loss | 0.781 | 0.796 |
| Autoencoder + KLDiv Loss | 0.859 | 0.825 |
Other networks
| Method | Clustering Accuracy | Normalized Mutual Information |
|---|---|---|
| DEC | 0.843 | 0.800 |
| DCN | 0.830 | 0.810 |
| CNN-RC | - | 0.915 |
| CNN-FD | - | 0.876 |
| DBC | 0.964 | 0.917 |
Note: The commit b34743114f68624b5371cd0d4c059b141422902f gives upto 0.96 accuracy and 0.92 NMI on the MNIST dataset. We will include it to the main branch once we can get better results with the COIL architecture
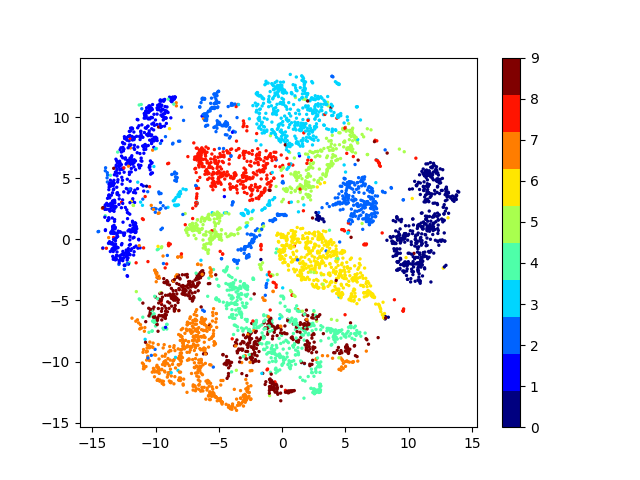
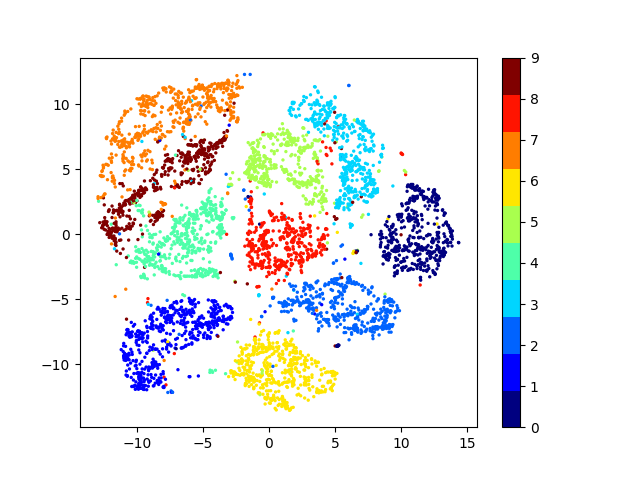
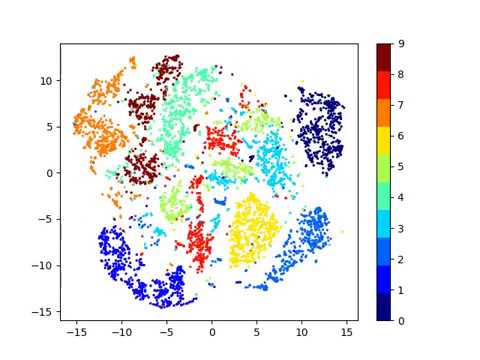
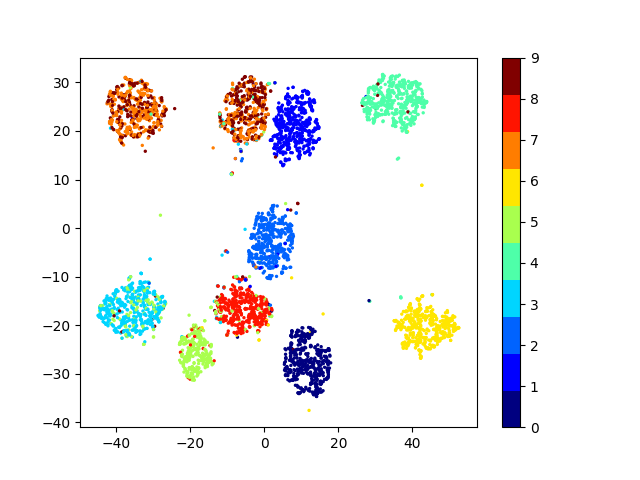
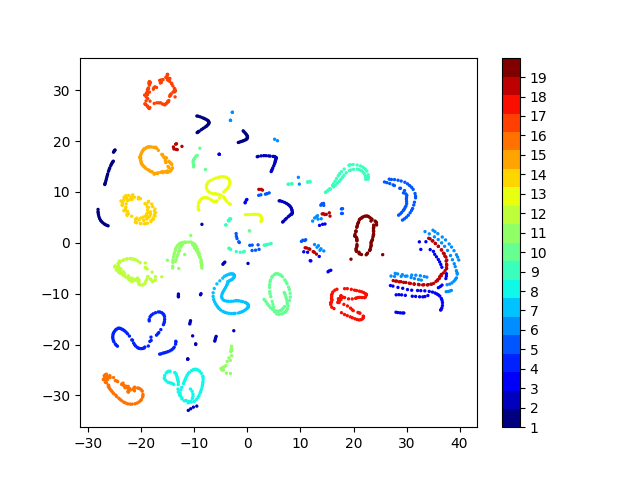
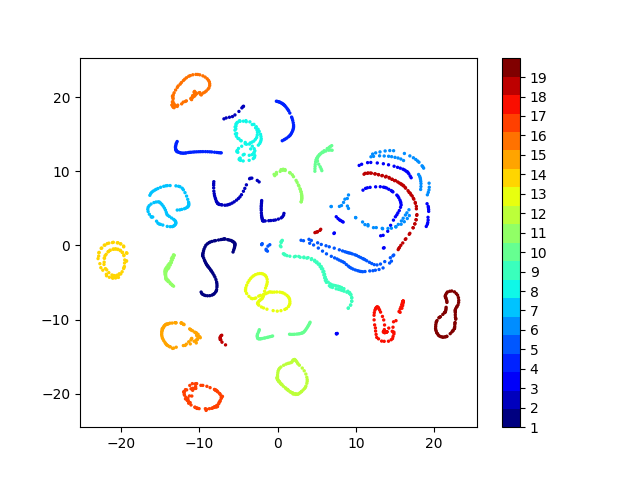
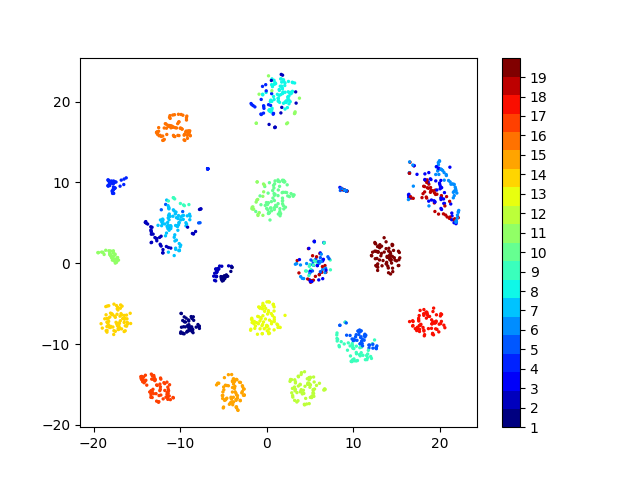
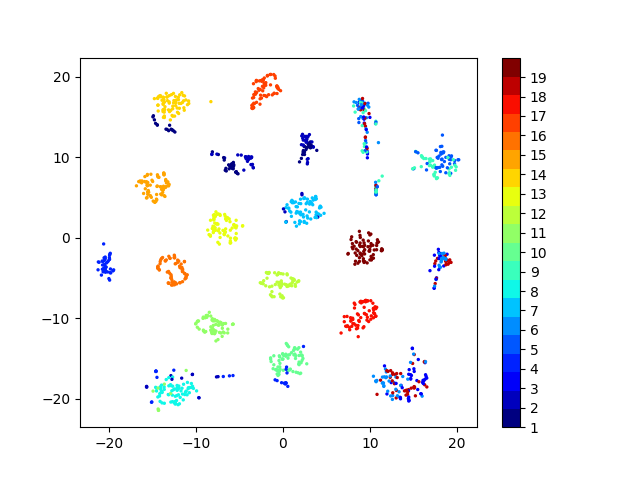
Latent space visualizations
Pixel space
Autoencoder
Autoencoder Latent Space Evolution (video)
Autoencoder + KLDivergence
Autoencoder + KLDivergence Latent Space Evolution (video)
Autoencoder + k-Means
COIL20
Our network
| Clustering space | Clustering Accuracy | Normalized Mutual Information |
|---|---|---|
| Image pixels | 0.689 | 0.793 |
| Autoencoder | 0.739 | 0.828 |
| Autoencoder + k-Means Loss | 0.745 | 0.846 |
| Autoencoder + KLDiv Loss | 0.762 | 0.848 |
Other networks
| Method | Clustering Accuracy | Normalized Mutual Information |
|---|---|---|
| DEN | 0.725 | 0.870 |
| CNN-RC | - | 1.000 |
| DBC | 0.793 | 0.895 |