cTPnet
single cell Transcriptome to Protein prediction with deep neural network (cTP-net) is a transfer learning framework to impute surface protein abundances from scRNA-seq data by learning from existing single-cell multi-omic resources. While single cell RNA sequencing (scRNA-seq) is invaluable for studying cell populations, cell-surface proteins are often integral markers of cellular function and serve as primary targets for therapeutic intervention.
Manuscript
Surface protein imputation from single cell transcriptomes by deep neural networks
Questions & Problems
If you have any questions or problems when using cTPnet or ctpnetpy, please feel free to open a new issue here. You can also email the maintainers of the corresponding packages -- the contact information is shown under Developers & Maintainers.
Pipeline overview
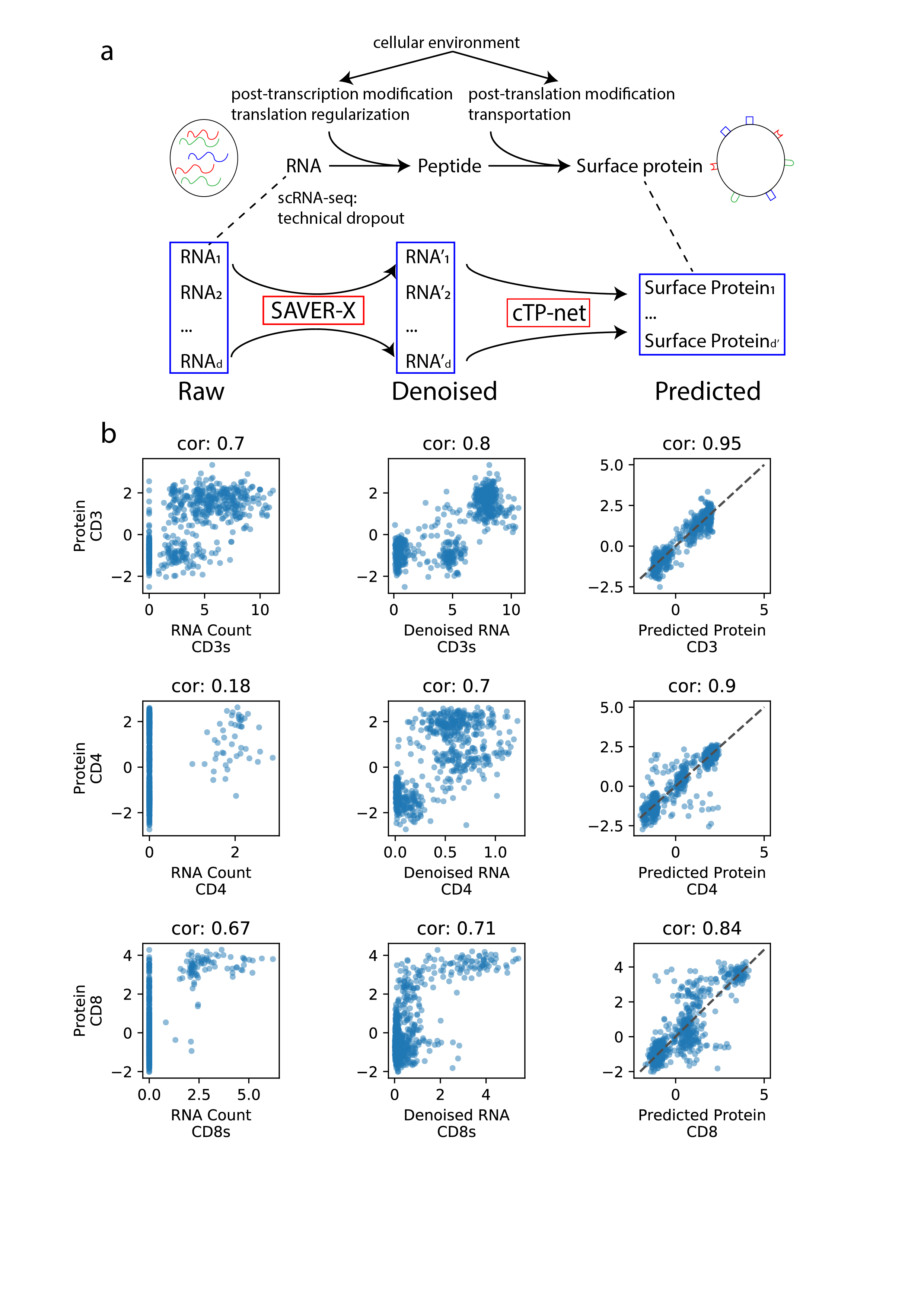
This cTPnet package includes two analysis tools: (1) SAVERX, we stronly recommend denoise the scRNA-seq data before impute the surface protein abundance, and (2) cTP-net, which impute the surface protein abundance based on previously trained model.
Figure 1. (a) Overview of cTP-net analysis pipeline, which learns a mapping from the denoised scRNA-seq data to the relative abundance of surface proteins, capturing multi-gene features that reflect the cellular environment and related processes. (b) For three example proteins, cross-cell scatter and correlation of CITE-seq measured abundances vs. (1) raw RNA count, (2) SAVER-X denoised RNA level, and (3) cTP-net predicted protein abundance.
Installation and running cTP-net
For different Seurat version, we developed separate vignette see below:
Seurat v2
cTP-net R notebook with step-by-step demonstration and rich display is available here. Corresponding Rmd script is available here.
Seurat v3
cTP-net R notebook with step-by-step demonstration and rich display is available here. Corresponding Rmd script is available here.
For a "normal" computer, the installation time is < 15mins. Given a dataset of 4000 cells, the running time of cTP-net should be less than 1 mins. No GPU required for running this algorithm.
Work in progress
As development in available cite-seq data set, we currently trainig a model that can extend our prediction to 100 surface proteins. Also, due to additional interets in training your own cTP-net model in the computational biology field, we have decided to share our training python code here. Please remember to cite our paper, and contacnt me or Nancy if for commercial usage.
Citation
Zhou, Z., Ye, C., Wang, J. et al. Surface protein imputation from single cell transcriptomes by deep neural networks. Nat Commun 11, 651 (2020). https://doi.org/10.1038/s41467-020-14391-0
Developers & Maintainers
-
Zilu Zhou (zhouzilu at pennmedicine dot upenn dot edu)
Genomics and Computational Biology Graduate Group, University of Pennsylvania -
Nancy R. Zhang (nzh at wharton dot upenn dot edu)
Department of Statistics, University of Pennsylvania
Common questions:
- Error message while running demo
Error in dim(X) <- c(n, length(X)/n) : dims [product 1992] do not match the length of object[2000]- Add
options(stringsAsFactors = FALSE)before running the code.
- Add
- Windows user if you see error message
Error in py_module_import(module, convert = convert) : ModuleNotFoundError: No module named 'ctpnet'- The solution is to use numpy 1.19.3.
pip install numpy==1.19.3 - For more information https://tinyurl.com/y3dm3h86
- In general, we don't recommend running cTP-net on a Windows platform even though I had some success at my intital submission of the code.
- The solution is to use numpy 1.19.3.