DolphinNext
A platform to create reproducible, portable and highly parallel pipelines
Citation: Yukselen, O., Turkyilmaz, O., Ozturk, A.R. et al. DolphinNext: a distributed data processing platform for high throughput genomics. BMC Genomics 21, 310 (2020). https://doi.org/10.1186/s12864-020-6714-x
DolphinNext, an intuitive web interface designed for users with limited bioinformatics experience to analyze and manage large numbers of samples on High Performance Computing (HPC) environments, cloud services or on a personal workstation.
- A platform to manage processing pipelines for large projects that require a scalable solution with automatic monitoring of large number of concurrent jobs
- A drag and drop user interface to create NextFlow pipelines.
- Run pipelines with different executors such as SGE, LSF, SLURM, Ignite etc.
Benefits of the design:
-
Build: Easily create new pipelines using a drag and drop interface. No need to write commands from scratch, instead reuse existing processes/modules to create new pipelines
-
Run: Execute pipelines in any host environment. Seamless Amazon Cloud and Google Cloud integration to create a cluster (instance), execute the pipeline and transfer the results to the storage services (S3 or GS).
-
Resume: A continuous checkpoint mechanism keeps track of each step of the running pipeline. Partially completed pipelines can be resumed at any stage even after parameter changes.
-
Improve: Revisioning system keeps track of pipelines and processes versions as well as their parameters. Edit, improve shared pipelines and customize them according to your needs.
-
Share: Share pipelines across different platforms. Isolate pipeline-specific dependencies in a container and easily replicate the methods in other clusters
Public Pipelines:
- RNA-Seq Pipelines (RSEM, HISAT, STAR, Tophat2)
- ATAC-Seq Pipeline
- ChIP Seq Pipeline
- Single Cell Pipelines (10X Genomics, Indrop)
- piRNA Pipelines (piPipes ChIP-Seq, Degradome/RAGE/CAGE, smallRNA)
- Sub Modules:
- Trimmer
- Adapter Removal
- Quality Filtering
- Common RNA Filtering
- ESAT
- FastQC,
- MultiQC
- RSeQC
- Picard
- IGV and UCSC genome browser file conversion
Overview:
Highlights:
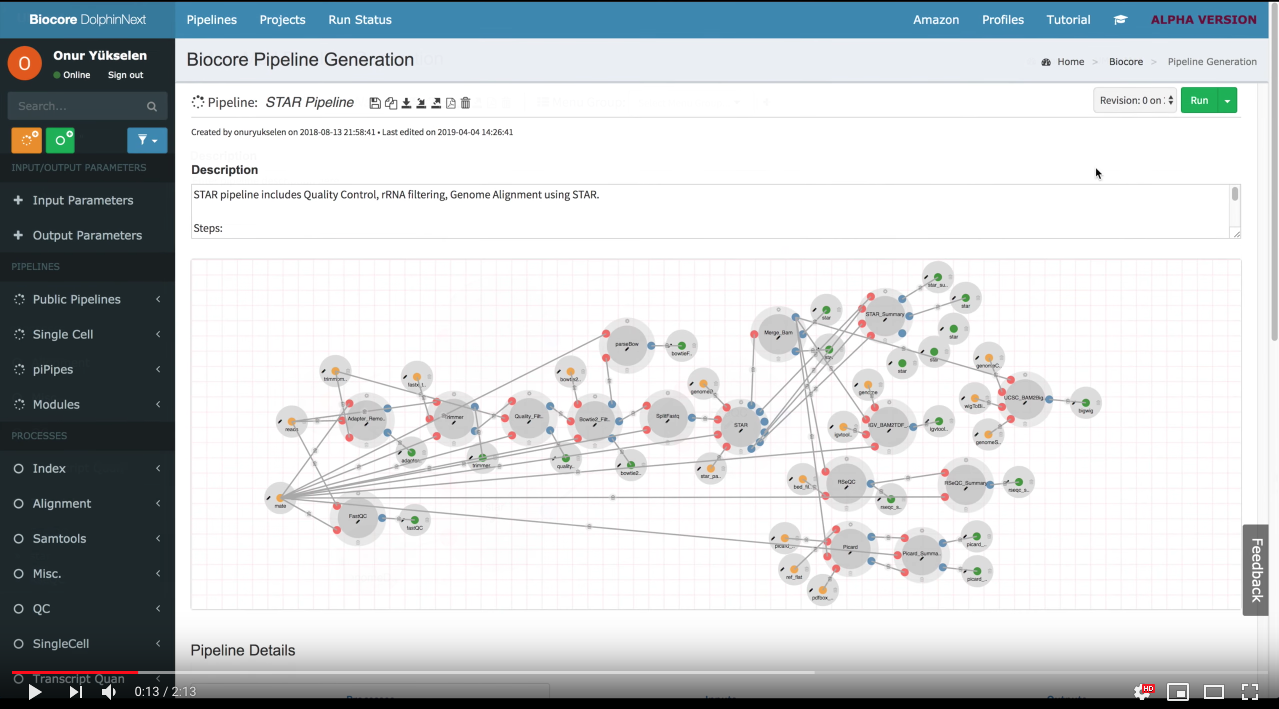
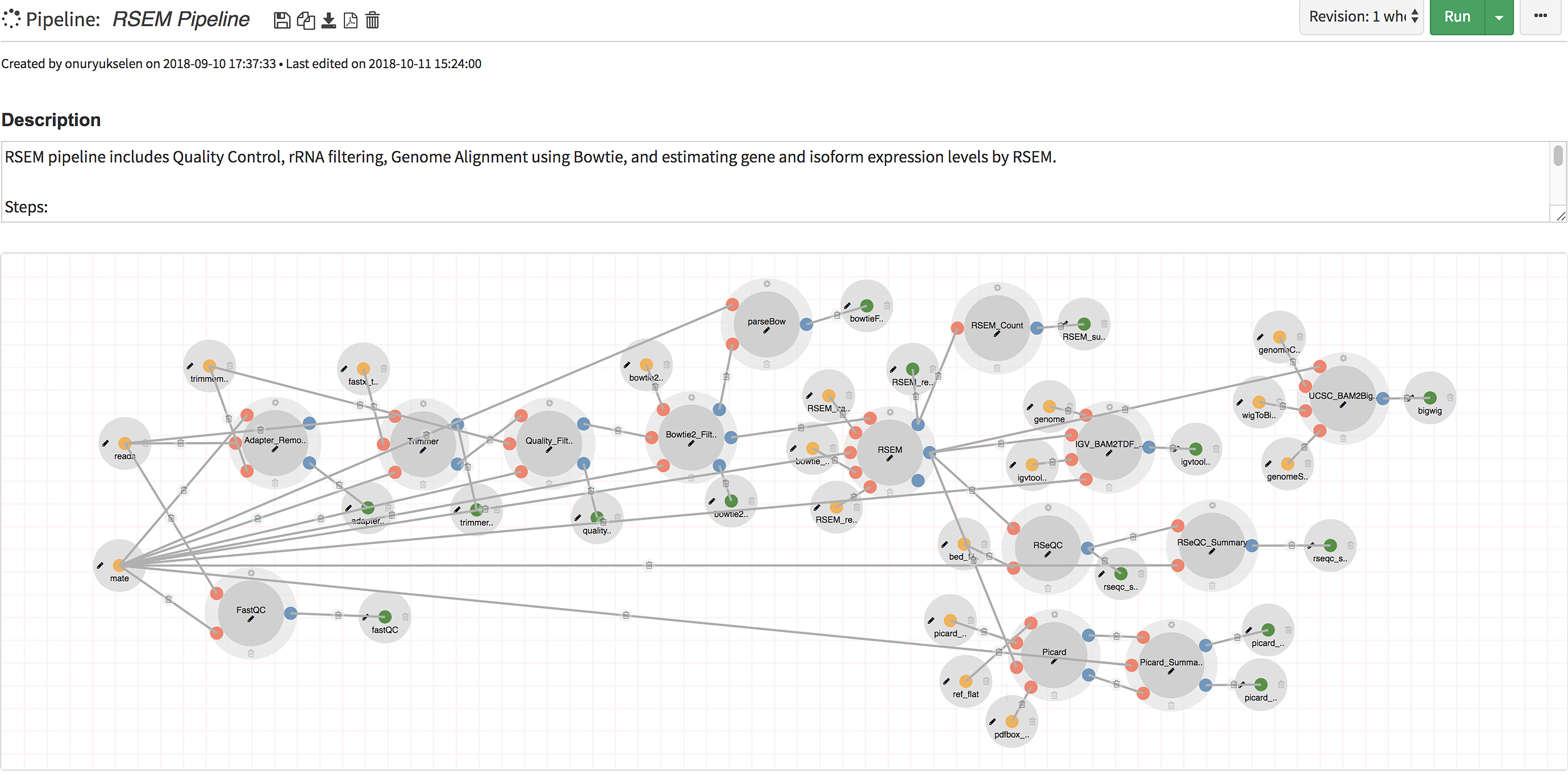
Workflow design with UI:
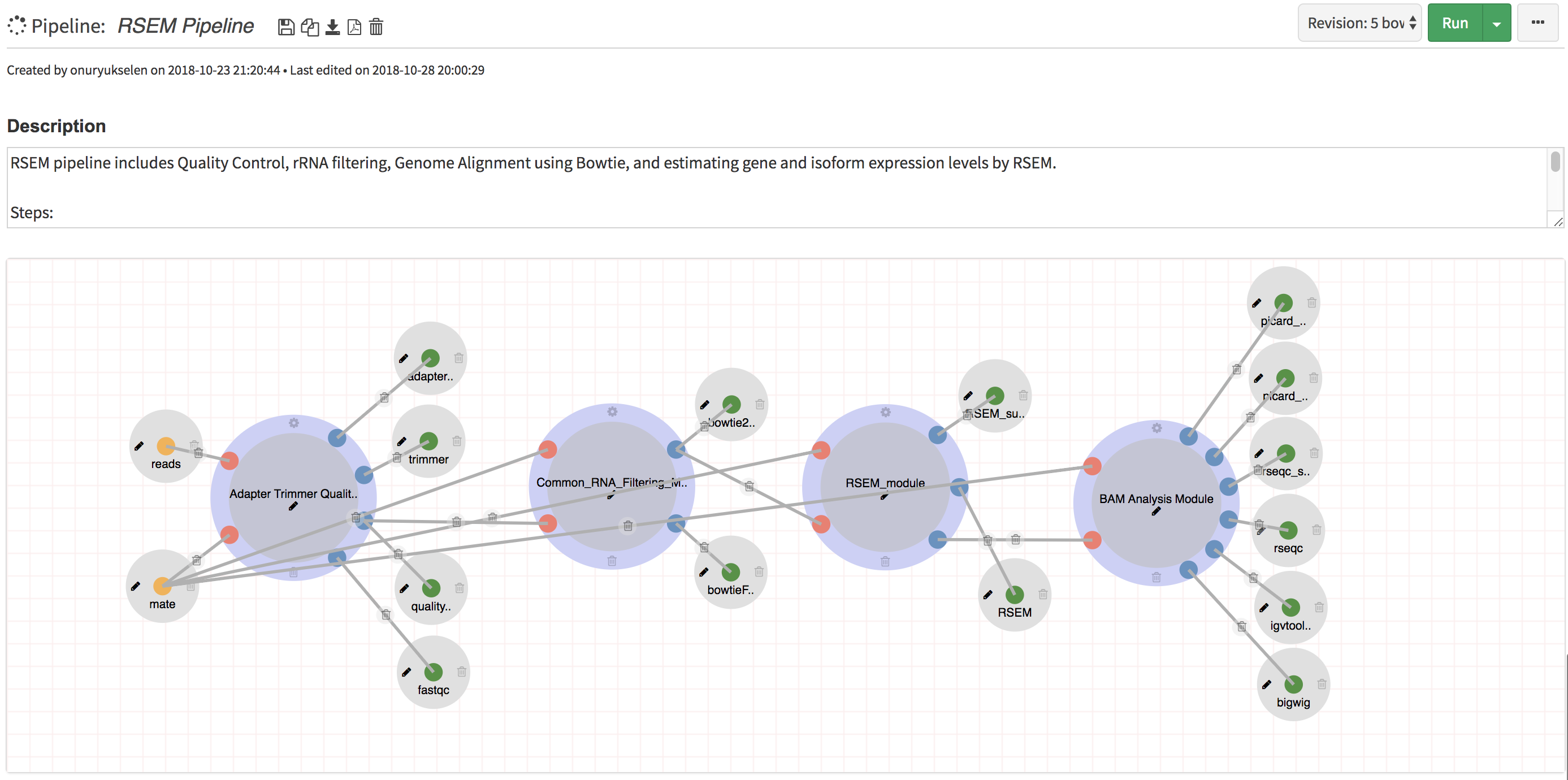
Modular System (Nested Workflows):
Quick Start and Documentation
Quick start guide, can be reached at https://dolphinnext.readthedocs.io/en/latest/dolphinNext/quick.html
Complete documentation is available at https://dolphinnext.readthedocs.io
Developer Tutorial is available which explains the basics of DolphinNext. You can use our website, or easily pull Docker image of Dolphinnext and start creating pipelines in your local server.
Citation:
If you use DolphinNext in your research, please cite:
Yukselen, O., Turkyilmaz, O., Ozturk, A.R. et al. DolphinNext: a distributed data processing platform for high throughput genomics. BMC Genomics 21, 310 (2020). https://doi.org/10.1186/s12864-020-6714-x
Support
UMMS Biocore, provides support for installations as well as commercial support for DolphinNext. Please contact [email protected]
Licensing
DolphinNext released under GNU General Public License 3.0.