briandconnelly / Growthcurve
Programming Languages
Labels
Projects that are alternatives of or similar to Growthcurve
growthcurve: Analysis of Biological Growth
The growthcurve package provides tools for analyzing biological growth, or "growth curves" in R. It is designed to integrate into modern workflows based around "tidy data", allowing it to be used in conjunction with other tools. growthcurve also provides wrappers that allow tidy growth data to be analyzed using the popular, but no-longer-maintained grofit package, if installed.
Installation
growthcurve is not quite ready to be available on CRAN, but you can use devtools to install the current development version:
if(!require("devtools")) install.packages("devtools")
devtools::install_github("briandconnelly/growthcurve", build_vignettes = TRUE)
Fitting Growth Curves
growthcurve's most important function is fit_growth, which fits a growth curve to the given data. Here, we'll fit a growth curve to one replicate population from the included pseudomonas data set, which has columns Time and CFUmL:
library(dplyr)
library(growthcurve)
rep1 <- filter(pseudomonas, Replicate == 1 & Strain == "PAO1")
myfit <- fit_growth(rep1, Time, CFUmL)
Even better, we can do this all at once with pipes:
myfit <- pseudomonas %>%
filter(Replicate == 1 & Strain == "PAO1") %>%
fit_growth(Time, CFUmL)
By default, fit_growth will fit a logistic curve, however the model argument can be used to specify a different model type. Here, we'll use a Gompertz function:
library(dplyr)
library(growthcurve)
rep1 <- filter(pseudomonas, Replicate == 1 & Strain == "PAO1")
myfit <- fit_growth(rep1, Time, CFUmL, model = "gompertz")
Other options include logistic4p, linear, loess, and spline. Additionally, grofit_logistic, grofit_gompertz, grofit_gompertz.exp, grofit_richards, and grofit_spline can be used to provide compatability with legacy scripts that use grofit. There's also a grofit_parametric, which finds the best among grofit's parametric models.
Interpreting Results
coming soon!
Visualizing Growth Curves
growthcurve includes tools for visualizing growth curves using either R's base graphics or ggplot2.
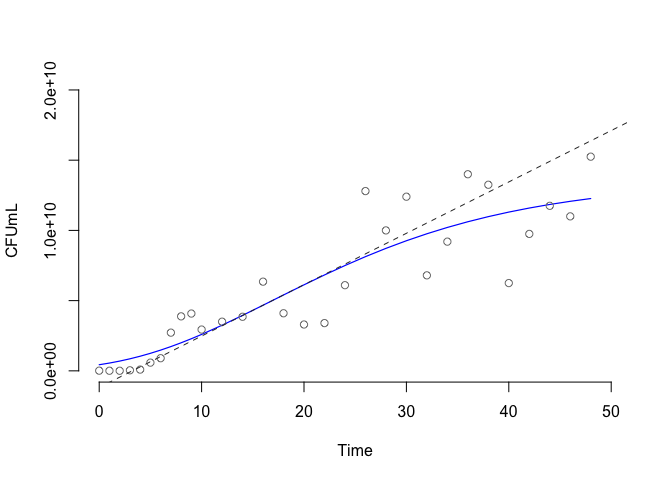
plot(myfit, show_raw = TRUE, show_maxrate = TRUE, show_asymptote = FALSE)
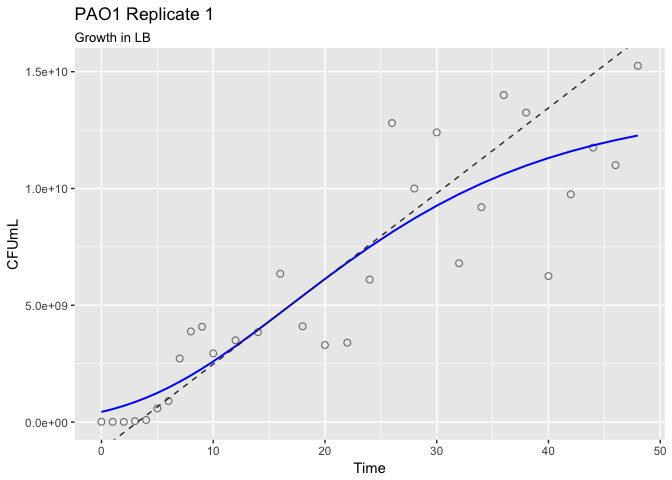
library(ggplot2)
autoplot(myfit, title = "PAO1 Replicate 1", subtitle = "Growth in LB")
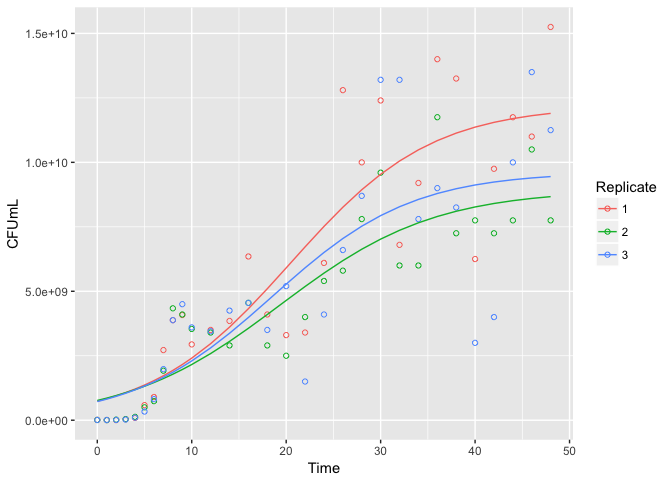
Alternatively, we can add growth curves to a ggplot2 plot with stat_growthcurve:
pao1data <- filter(pseudomonas, Strain == "PAO1")
ggplot(data = pao1data, aes(x = Time, y = CFUmL, color = Replicate)) +
geom_point(shape = 1) +
stat_growthcurve()
Code of Conduct
This project is released with a Contributor Code of Conduct. By participating in this project, you agree to abide by its terms.
License
growthcurve is released under the Simplified BSD License.