MrGiovanni / Modelsgenesis
Projects that are alternatives of or similar to Modelsgenesis
We have built a set of pre-trained models called Generic Autodidactic Models, nicknamed Models Genesis, because they are created ex nihilo (with no manual labeling), self-taught (learned by self-supervision), and generic (served as source models for generating application-specific target models). We envision that Models Genesis may serve as a primary source of transfer learning for 3D medical imaging applications, in particular, with limited annotated data.
Paper
This repository provides the official implementation of training Models Genesis as well as the usage of the pre-trained Models Genesis in the following paper:
Models Genesis: Generic Autodidactic Models for 3D Medical Image Analysis
Zongwei Zhou1, Vatsal Sodha1, Md Mahfuzur Rahman Siddiquee1,
Ruibin Feng1, Nima Tajbakhsh1, Michael B. Gotway2, and Jianming Liang1
1 Arizona State University, 2 Mayo Clinic
International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2019
Young Scientist Award
paper | code | slides | poster | talk (YouTube, YouKu) | blog
Models Genesis
Zongwei Zhou1, Vatsal Sodha1, Jiaxuan Pang1, Michael B. Gotway2, and Jianming Liang1
1 Arizona State University, 2 Mayo Clinic
Medical Image Analysis (MedIA)
MedIA Best Paper Award
paper | code | slides
Available implementation
- keras/
- pytorch/
★ News: Models Genesis, incorporated with nnU-Net, rank # 1 in segmenting liver/tumor and hippocampus.
- competition/
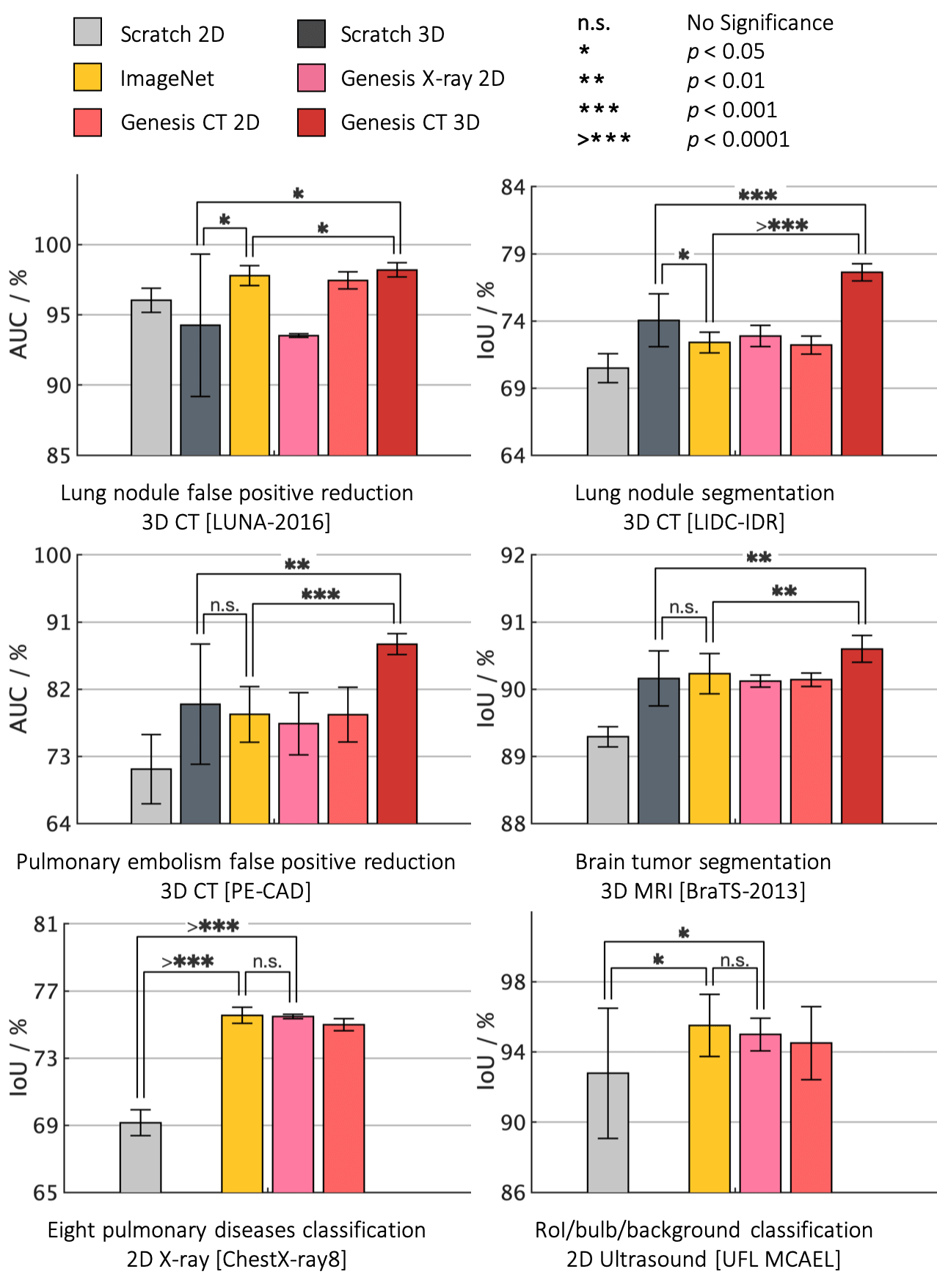
Major results from our work
- Models Genesis outperform 3D models trained from scratch
- Models Genesis top any 2D approaches, including ImageNet models and degraded 2D Models Genesis
- Models Genesis (2D) offer performances equivalent to supervised pre-trained models
The par plots presented below are produced by Matlab code in figures/plotsuperbar.m and the helper functions in figures/superbar. Credit to superbar by Scott Lowe.
Note that learning from scratch simply in 3D may not necessarily yield performance better than ImageNet-based transfer learning in 2D
Citation
If you use this code or use our pre-trained weights for your research, please cite our papers:
@InProceedings{zhou2019models,
author="Zhou, Zongwei and Sodha, Vatsal and Rahman Siddiquee, Md Mahfuzur and Feng, Ruibin and Tajbakhsh, Nima and Gotway, Michael B. and Liang, Jianming",
title="Models Genesis: Generic Autodidactic Models for 3D Medical Image Analysis",
booktitle="Medical Image Computing and Computer Assisted Intervention -- MICCAI 2019",
year="2019",
publisher="Springer International Publishing",
address="Cham",
pages="384--393",
isbn="978-3-030-32251-9",
url="https://link.springer.com/chapter/10.1007/978-3-030-32251-9_42"
}
@article{zhou2021models,
title="Models Genesis",
author="Zhou, Zongwei and Sodha, Vatsal and Pang, Jiaxuan and Gotway, Michael B and Liang, Jianming",
journal="Medical Image Analysis",
volume = "67",
pages = "101840",
year = "2021",
issn = "1361-8415",
doi = "https://doi.org/10.1016/j.media.2020.101840",
url = "http://www.sciencedirect.com/science/article/pii/S1361841520302048",
}
Acknowledgement
This research has been supported partially by ASU and Mayo Clinic through a Seed Grant and an Innovation Grant, and partially by the National Institutes of Health (NIH) under Award Number R01HL128785. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work has utilized the GPUs provided partially by the ASU Research Computing and partially by the Extreme Science and Engineering Discovery Environment (XSEDE) funded by the National Science Foundation (NSF) under grant number ACI-1548562. This is a patent-pending technology.