openworm / Owmeta
Programming Languages
Projects that are alternatives of or similar to Owmeta
owmeta
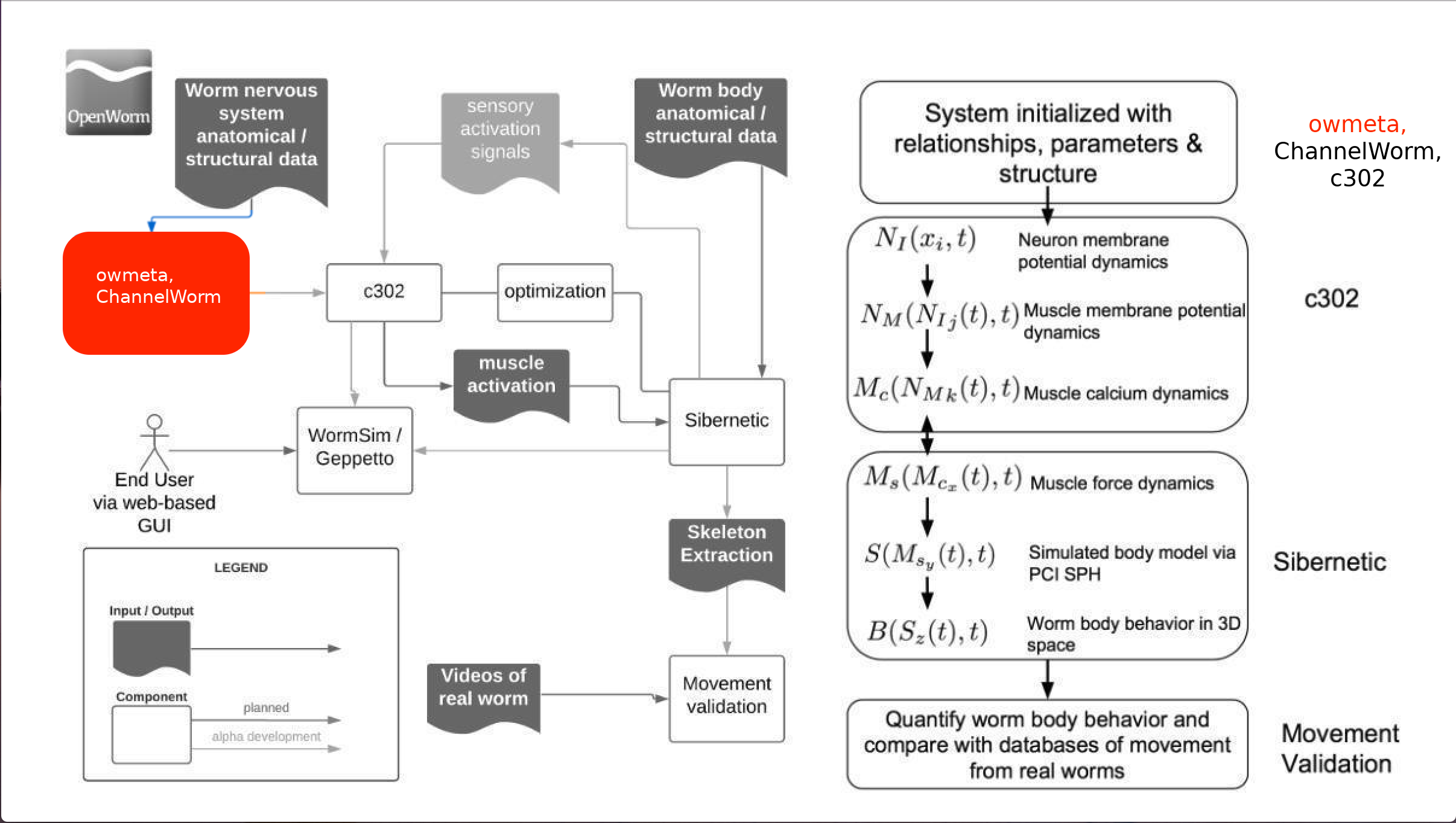
A data access layer in Python which integrates disparate structures and representations for C. elegans anatomy and physiology. Enables a simple Python API for asking various questions about the cells of the C. elegans and enabling data sharing for the purpose of building a data-to-model pipeline for the OpenWorm project.
Overview
The data and models required to simulate C. elegans are highly heterogeneous. Consequently, from a software perspective, a variety of underlying representations are needed to store different aspects of the relevant anatomy and physiology. For example, a NetworkX representation of the connectome as a complex graph enables questions to be asked about nearest neighbors of a given neuron. An RDF semantic graph representation is useful for reading and writing annotations about multiple aspects of a neuron, such as what papers have been written about it, properties it may have such as ion channels and neurotransmitter receptors, etc. A NeuroML representation is useful for answering questions about model morphology and simulation parameters. A Blender representation is a full 3D shape definition that can be used for calculations in 3D space.
The diversity of underlying representations required for OpenWorm presents a challenge for data integration and consolidation. owmeta solves this challenge with a unified data access layer whereby different representations are encapsulated into an abstract view. This allows the user to work with objects related to the biological reality of the worm, and forget about which representation is being used under the hood. The worm itself has a unified sense of neurons, networks, muscles, ion channels, etc. and so should our code.
Relationship to ChannelWorm2
ChannelWorm2 is the sub-project of OpenWorm which houses ion channel models. In the future, we expect ChannelWorm2 to be a "consumer" of owmeta. An owmeta database will house physical models, the digitized plots they are derived from (there is a Plot type in owmeta), and provide code to put those models into enumerated formats along with auxiliary files or comments. However, because these projects were not developed sequentially, there is currently some overlap in functionality, and owmeta itself houses a fairly substantial amount of physiological information about C. elegans. Ultimately, the pure core of owmeta, which is meant to be a data framework for storing metadata and provenance (i.e. parameters and trajectories associated with simulations), will be separated out into standalone functionality.
Versioning data as code
A library that attempts to reliably expose dynamic data can often be broken because the underlying data sets that define it change over time. This is because data changes can cause queries to return different answers than before, causing unpredictable behavior.
As such, to create a stable foundational library for others to reuse, the version of the owmeta library guarantees the user a specific version of the data behind that library. In addition, unit tests are used to ensure basic sanity checks on data are maintained. As data are improved, the maintainers of the library can perform appropriate regression tests prior to each new release to guarantee stability.
Installation
See INSTALL.md
Quickstart
To get started, you'll need to connect to a database. The OpenWorm owmeta
"project" is currently hosted at https://github.com/openworm/OpenWormData.git.
This project holds a working-copy of the database. You can retrieve it by
executing the following command line after owmeta installation:
owm clone https://github.com/openworm/OpenWormData.git --branch owmeta
This command should create a directory .owm in your current working
directory. Then, in Python, from the same working directory:
>>> from owmeta_core.command import OWM
>>> conn = OWM().connect()
This creates a connection to the project stored under the .owm directory.
Then you can try out a few things:
# Make the context
>>> from owmeta_core.context import Context
>>> ctx = conn(Context)(ident='http://openworm.org/data')
# Grab the representation of the neuronal network
>>> from owmeta.worm import Worm
>>> net = ctx.stored(Worm).query().neuron_network()
# Grab a specific neuron
>>> from owmeta.neuron import Neuron
>>> aval = ctx.stored(Neuron).query(name='AVAL')
# Get the neuron's type
>>> aval.type.one()
'interneuron'
# Count how many connections come from AVAL
>>> aval.connection.count('pre')
86
More examples
Return information about individual neurons:
>>> aval.name()
'AVAL'
# List all known receptors
>>> sorted(aval.receptors())
['GGR-3', 'GLR-1', ... 'NPR-4', 'UNC-8']
# Show how many chemical synapses go in and out of AVAL
>>> aval.connection.count('either', syntype='send')
105
Return the list of all neurons:
>>> len(set(net.neuron_names()))
302
>>> sorted(net.neuron_names())
['ADAL', 'ADAR', ... 'VD8', 'VD9']
Return a set of all muscles:
>>> muscles = ctx.stored(Worm).query().muscles()
>>> len(muscles)
158
Because the ultimate aim of OpenWorm is to be a platform for biological research, the physiological data in owmeta should be uncontroversial and well supported by evidence. Using the Evidence type, it is possible to link data and models to corresponding articles from peer-reviewed literature:
>>> from owmeta.document import Document
>>> from owmeta.evidence import Evidence
# Make a context for evidence (i.e., statements about other groups of statements)
>>> evctx = conn(Context)(ident='http://example.org/evidence/context')
# Make a context for defining domain knowledge
>>> dctx = conn(Context)(ident='http://example.org/data/context')
>>> doc = evctx(Document)(key="Sulston83", author='Sulston et al.', date='1983')
>>> e = evctx(Evidence)(key="Sulston83", reference=doc)
>>> avdl = dctx(Neuron)(name="AVDL")
>>> avdl.lineageName("AB alaaapalr")
owmeta_core.statement.Statement(subj=Neuron(ident=rdflib.term.URIRef('http://data.openworm.org/sci/bio/Neuron#AVDL')), prop=owmeta.cell.Cell_lineageName(owner=Neuron(ident=rdflib.term.URIRef('http://data.openworm.org/sci/bio/Neuron#AVDL'))), obj=owmeta_core.dataobject_property.ContextualizedPropertyValue(rdflib.term.Literal('AB alaaapalr')), context=owmeta_core.context.Context(ident="http://example.org/data/context"))
>>> e.supports(dctx.rdf_object)
owmeta_core.statement.Statement(subj=Evidence(ident=rdflib.term.URIRef('http://data.openworm.org/Evidence#Sulston83')), prop=owmeta.evidence.Evidence_supports(owner=Evidence(ident=rdflib.term.URIRef('http://data.openworm.org/Evidence#Sulston83'))), obj=ContextDataObject(ident=rdflib.term.URIRef('http://example.org/data/context')), context=owmeta_core.context.Context(ident="http://example.org/evidence/context"))
>>> dctx.save_context()
>>> evctx.save_context()
Retrieve evidence:
>>> doc = evctx.stored(Document)(author='Sulston et al.', date='1983')
>>> e0 = evctx.stored(Evidence)(reference=doc)
>>> supported_ctx = e0.supports()
# is the neuron's presence asserted?
>>> dctx.identifier == supported_ctx.identifier
True
Query for neurons in C. elegans:
>>> from owmeta.network import Network
# The default Worm() is for C. elegans
>>> with ctx.stored(Worm, Neuron, Network) as cctx:
... w = cctx.Worm()
... net = cctx.Network()
... w.neuron_network(net)
owmeta_core.statement.Statement(subj=Worm(ident=rdflib.term.URIRef('http://data.openworm.org/sci/bio/Worm#a8020ed8519038a6bbc98f1792c46c97b')), prop=owmeta.worm.Worm_neuron_network(owner=Worm(ident=rdflib.term.URIRef('http://data.openworm.org/sci/bio/Worm#a8020ed8519038a6bbc98f1792c46c97b'))), obj=Network(ident=rdflib.term.URIRef('http://data.openworm.org/sci/bio/Network#a5859bb1e51537f60e506c283401fcd84')), context=owmeta_core.context.QueryContext(ident="http://openworm.org/data"))
... neur = cctx.Neuron.query()
... net.neuron(neur)
... neur.count()
302
See what neurons express a given neuropeptide:
>>> n = ctx.stored(Neuron).query()
>>> n.neuropeptide("INS-26")
owmeta_core.statement.Statement(subj=Neuron(ident=rdflib.term.Variable('aNeuron_...')), prop=owmeta.neuron.Neuron_neuropeptide(owner=Neuron(ident=rdflib.term.Variable('aNeuron_...'))), obj=owmeta_core.dataobject_property.ContextualizedPropertyValue(rdflib.term.Literal('INS-26')), context=owmeta_core.context.QueryContext(ident="http://openworm.org/data"))
>>> sorted(x.name() for x in n.load())
['ASEL', 'ASER', 'ASIL', 'ASIR']
Get direct access to the RDFLib graph:
>>> conn.rdf.query("SELECT ?y WHERE { ?x rdf:type ?y }")
<rdflib.plugins.sparql.processor.SPARQLResult object at ...>
Modeling data
As described above, ultimately, ion channel models will be part of the ChannelWorm2 repository. As the project evolves, other models, such as for reproduction and development, may be housed in their own repositories. But for the time being, the owmeta repository contains specific models as well. These models will eventually be transferred to an appropriate and independent data repository within the OpenWorm suite of tools.
# Get data for a subtype of voltage-gated potassium channels
>>> from owmeta.channel import Channel
>>> kv1 = ctx(Channel)(subfamily='Kv1.1')
>>> kv1.models()
The same type of operation can be used to obtain the experimental data a given model was derived from.
# Get experiment(s) that back up the data model
>> some_model = mods[0]
>> some_model.references.get()
Finally, when you're done accessing the database, be sure to disconnect from it:
>>> conn.disconnect()
More examples can be found here and here.
Documentation
Further documentation is available online.
Contributing
We happily welcome pull requests and bug reports. If, you are not sure how you can contribute, fill out this (short) form, and you'll receive an invite to our Slack chat where you can initiate more in-depth conversations.
Questions/Concerns?
You can ask questions, leave bug reports, or propose features on our issue tracker.

