PumasAI / Pumas.jl
Programming Languages
Projects that are alternatives of or similar to Pumas.jl
Pumas.jl
Pumas: A Pharmaceutical Modeling and Simulation toolkit
Resources
Demo: A Simple PK model
using Pumas, Plots
For reproducibility, we will set a random seed:
using Random
Random.seed!(1)
A simple one compartment oral absorption model using an analytical solution
model = @model begin
@param begin
tvcl ∈ RealDomain(lower=0)
tvv ∈ RealDomain(lower=0)
pmoncl ∈ RealDomain(lower = -0.99)
Ω ∈ PDiagDomain(2)
σ_prop ∈ RealDomain(lower=0)
end
@random begin
η ~ MvNormal(Ω)
end
@covariates wt isPM
@pre begin
CL = tvcl * (1 + pmoncl*isPM) * (wt/70)^0.75 * exp(η[1])
V = tvv * (wt/70) * exp(η[2])
end
@dynamics Central1
#@dynamics begin
# Central' = - (CL/V)*Central
#end
@derived begin
cp = @. 1000*(Central / V)
dv ~ @. Normal(cp, sqrt(cp^2*σ_prop))
end
end
Develop a simple dosing regimen for a subject
ev = DosageRegimen(100, time=0, addl=4, ii=24)
s1 = Subject(id=1, evs=ev, cvs=(isPM=1, wt=70))
Simulate a plasma concentration time profile
param = (
tvcl = 4.0,
tvv = 70,
pmoncl = -0.7,
Ω = Diagonal([0.09,0.09]),
σ_prop = 0.04
)
obs = simobs(model, s1, param, obstimes=0:1:120)
plot(obs)
Generate a population of subjects
choose_covariates() = (isPM = rand([1, 0]),
wt = rand(55:80))
pop_with_covariates = Population(map(i -> Subject(id=i, evs=ev, cvs=choose_covariates()),1:10))
Simulate into the population
obs = simobs(model, pop_with_covariates, param, obstimes=0:1:120)
and visualize the output
plot(obs)
Let's roundtrip this simulation to test our estimation routines
simdf = DataFrame(obs)
simdf.cmt .= 1
first(simdf, 6)
Read the data in to Pumas
data = read_pumas(simdf, time=:time,cvs=[:isPM, :wt])
Evaluating the results of a model fit goes through an fit --> infer --> inspect --> validate cycle
fit
julia> res = fit(model,data,param,Pumas.FOCEI())
FittedPumasModel
Successful minimization: true
Likelihood approximation: Pumas.FOCEI
Objective function value: 8363.13
Total number of observation records: 1210
Number of active observation records: 1210
Number of subjects: 10
-----------------
Estimate
-----------------
tvcl 4.7374
tvv 68.749
pmoncl -0.76408
Ω₁,₁ 0.046677
Ω₂,₂ 0.024126
σ_prop 0.041206
-----------------
infer
infer provides the model inference
julia> infer(res)
Calculating: variance-covariance matrix. Done.
FittedPumasModelInference
Successful minimization: true
Likelihood approximation: Pumas.FOCEI
Objective function value: 8363.13
Total number of observation records: 1210
Number of active observation records: 1210
Number of subjects: 10
---------------------------------------------------
Estimate RSE 95.0% C.I.
---------------------------------------------------
tvcl 4.7374 10.057 [ 3.8036 ; 5.6713 ]
tvv 68.749 5.013 [61.994 ; 75.503 ]
pmoncl -0.76408 -3.9629 [-0.82342 ; -0.70473 ]
Ω₁,₁ 0.046677 34.518 [ 0.015098 ; 0.078256]
Ω₂,₂ 0.024126 31.967 [ 0.0090104; 0.039243]
σ_prop 0.041206 3.0853 [ 0.038714 ; 0.043698]
---------------------------------------------------
inspect
inspect gives you the model predictions, residuals and Empirical Bayes estimates
resout = DataFrame(inspect(res))
julia> first(resout, 6)
6×13 DataFrame
│ Row │ id │ time │ isPM │ wt │ pred │ ipred │ pred_approx │ wres │ iwres │ wres_approx │ ebe_1 │ ebe_2 │ ebes_approx │
│ │ String │ Float64 │ Int64 │ Int64 │ Float64 │ Float64 │ Pumas.FOCEI │ Float64 │ Float64 │ Pumas.FOCEI │ Float64 │ Float64 │ Pumas.FOCEI │
├─────┼────────┼─────────┼───────┼───────┼─────────┼─────────┼─────────────┼──────────┼───────────┼─────────────┼───────────┼───────────┼─────────────┤
│ 1 │ 1 │ 0.0 │ 1 │ 74 │ 1344.63 │ 1679.77 │ FOCEI() │ 0.273454 │ -0.638544 │ FOCEI() │ -0.189025 │ -0.199515 │ FOCEI() │
│ 2 │ 1 │ 0.0 │ 1 │ 74 │ 1344.63 │ 1679.77 │ FOCEI() │ 0.273454 │ -0.638544 │ FOCEI() │ -0.189025 │ -0.199515 │ FOCEI() │
│ 3 │ 1 │ 0.0 │ 1 │ 74 │ 1344.63 │ 1679.77 │ FOCEI() │ 0.273454 │ -0.638544 │ FOCEI() │ -0.189025 │ -0.199515 │ FOCEI() │
│ 4 │ 1 │ 0.0 │ 1 │ 74 │ 1344.63 │ 1679.77 │ FOCEI() │ 0.273454 │ -0.638544 │ FOCEI() │ -0.189025 │ -0.199515 │ FOCEI() │
│ 5 │ 1 │ 0.0 │ 1 │ 74 │ 1344.63 │ 1679.77 │ FOCEI() │ 0.273454 │ -0.638544 │ FOCEI() │ -0.189025 │ -0.199515 │ FOCEI() │
│ 6 │ 1 │ 0.0 │ 1 │ 74 │ 1344.63 │ 1679.77 │ FOCEI() │ 0.273454 │ -0.638544 │ FOCEI() │ -0.189025 │ -0.199515 │ FOCEI() │
Simulate from fitted model
In order to simulate from a fitted model simobs can be used. The final parameters of the fitted models are available in the coef(res)
fitparam = coef(res)
You can then pass these optimized parameters into a simobs call and pass the same dataset or simulate into a different design
ev_sd_high_dose = DosageRegimen(200, time=0, addl=4, ii=48)
s2 = Subject(id=1, evs=ev_sd_high_dose, cvs=(isPM=1, wt=70))
obs = simobs(model, s2, fitparam, obstimes=0:1:160)
plot(obs)
Visualization
There are several plot recipes you can use to visualize your model fit. These are mainly provided by the PumasPlots.jl package (currently unregistered, add via URL).
PumasPlots provides recipes to visualize FittedPumasModels, and also has powerful grouping functionality. While some plot types are specialized for fitted models, you can use all of Plots' features by converting your result to a DataFrame (using DataFrame(inspect(res))).
-
convergence(res::FittedPumasModel)- plots the optimization trajectory of all variables.convergence(res)
-
etacov(res::FittedPumasModel)- plots the covariates of the model against the etas. Keyword arguments are described in the docstring - essentially, you can use any column inDataFrame(inspect(res))to plot.etacov(res; catmap = (isPM = true,))
-
resplot(res::FittedPumasModel)- plots the covariates of the model against their residuals, determined by the approximation type. Accepts many of the same kwargs asetacov.resplot(res; catmap = (isPM = true,))
-
corrplot(empirical_bayes(res); labels)- overload of theStatsPlotscorrplot, for etas.corrplot(empirical_bayes(res); labels = ["eta_1", "eta_2"])
Most of these plotting functions have docstrings, which can be accessed in the REPL help mode by >? resplot (for example).
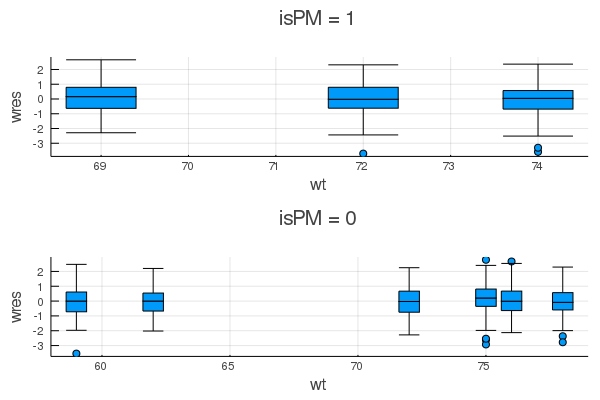
In addition to these specialized plots, PumasPlots offers powerful grouping functionality. By working with the DataFrame of your results (DataFrame(inspect(res))), you can create arbitrary plots, and by using the plot_grouped function, you can create panelled (a.k.a. grouped) plots.
df = DataFrame(inspect(res))
gdf = groupby(df, :isPM) # group by categorical covariate
plot_grouped(gdf) do subdf # `plot_grouped` will iterate through the groups of `gdf`
boxplot(subdf.wt, subdf.wres; xlabel = :wt, ylabel = :wres, legend = :none) # you can use any arbitrary plotting function here
end