biocore-ntnu / Pyranges
Programming Languages
Projects that are alternatives of or similar to Pyranges
pyranges
Introduction
GenomicRanges and genomic Rle-objects for Python.
"Finally ... This was what Python badly needed for years." - Heng Li
Release
PyRanges is in a beta state until any possible issues with the move to pandas 1.0 have been ironed out. We are extremely responsive to bug-reports, so if you have problems or come across unexpected behavior, please create an issue.
Asking for help
Feel free to ask questions of the type "how do I do X with pyranges?" on public forums like Stack Overflow, Bioinformatics Stack Exchange or Biostars. You can use endrebak85#gmail.com to point me to the question(s).
Quick examples
import pyranges as pr
# load example datasets
exons, cpg = pr.data.exons(), pr.data.cpg()
# subsetting pyranges is easy
exons["chrY", "-", 15591259:27197945]
# +--------------|-----------|-----------|----------------------------------------|-----------|--------------+
# | Chromosome | Start | End | Name | Score | Strand |
# | (category) | (int32) | (int32) | (object) | (int64) | (category) |
# |--------------|-----------|-----------|----------------------------------------|-----------|--------------|
# | chrY | 15591393 | 15592550 | NR_047610_exon_27_0_chrY_15591394_r | 0 | - |
# | chrY | 15591393 | 15592550 | NR_047607_exon_29_0_chrY_15591394_r | 0 | - |
# | chrY | 15591393 | 15592550 | NM_001258269_exon_29_0_chrY_15591394_r | 0 | - |
# | chrY | 15591393 | 15592550 | NR_047599_exon_28_0_chrY_15591394_r | 0 | - |
# | ... | ... | ... | ... | ... | ... |
# | chrY | 25336491 | 25336631 | NM_004081_exon_22_0_chrY_25336492_r | 0 | - |
# | chrY | 26952215 | 26952307 | NM_020364_exon_16_0_chrY_26952216_r | 0 | - |
# | chrY | 27197822 | 27197945 | NM_004678_exon_7_0_chrY_27197823_r | 0 | - |
# | chrY | 27197822 | 27197945 | NM_001002760_exon_7_0_chrY_27197823_r | 0 | - |
# +--------------|-----------|-----------|----------------------------------------|-----------|--------------+
# Stranded PyRanges object has 22 rows and 6 columns from 1 chromosomes.
# you can use your pandas-skills with pyranges
exons[~exons.Name.str.startswith("NR")] # all rows where the name column does not start with "NR"
# +--------------|-----------|-----------|----------------------------------------|-----------|--------------+
# | Chromosome | Start | End | Name | Score | Strand |
# | (category) | (int32) | (int32) | (object) | (int64) | (category) |
# |--------------|-----------|-----------|----------------------------------------|-----------|--------------|
# | chrX | 135574120 | 135574598 | NM_001727_exon_2_0_chrX_135574121_f | 0 | + |
# | chrX | 47868945 | 47869126 | NM_205856_exon_4_0_chrX_47868946_f | 0 | + |
# | chrX | 77294333 | 77294480 | NM_000052_exon_17_0_chrX_77294334_f | 0 | + |
# | chrX | 91090459 | 91091043 | NM_001168360_exon_0_0_chrX_91090460_f | 0 | + |
# | ... | ... | ... | ... | ... | ... |
# | chrY | 15481135 | 15481229 | NM_182659_exon_16_0_chrY_15481136_r | 0 | - |
# | chrY | 25325872 | 25325936 | NM_004081_exon_18_0_chrY_25325873_r | 0 | - |
# | chrY | 15560896 | 15560946 | NM_001258258_exon_25_0_chrY_15560897_r | 0 | - |
# | chrY | 15467254 | 15467278 | NM_001258270_exon_13_0_chrY_15467255_r | 0 | - |
# +--------------|-----------|-----------|----------------------------------------|-----------|--------------+
# Stranded PyRanges object has 847 rows and 6 columns from 2 chromosomes.
# the API allows for easy and terse chaining
(cpg # use the cpg dataset
.join(exons, suffix="_xn") # join with exons, use suffix _xn for duplicate cols
.subset(lambda df: df.CpG > 30) # keep only rows with a CpG score over 30
.sort(nb_cpu=2) # sort on Chromosome, Start and End
# note that virtually all pyranges-methods take a nb_cpu argument
# to use multiple cores, you need to install ray with pip install ray
.pc(formatting={"Start": "{:,}", "End": "{:,}", "Name": "{:2.2}"})
# print, while keeping the chain (c) going. Try .sp(), msp(), rp(), spc(), mspc() also :)
["chrX"] # keep only chromosome X
.assign("CpGDecile", lambda df: df.CpG / 10) # Insert new column
.unstrand()) # remove the strand info
# +--------------|-------------|-------------|-----------|------------|-----------|------------|-----------|--------------+
# | Chromosome | Start | End | CpG | Start_xn | End_xn | Name | Score | Strand |
# | (category) | (int32) | (int32) | (int64) | (int32) | (int32) | (object) | (int64) | (category) |
# |--------------|-------------|-------------|-----------|------------|-----------|------------|-----------|--------------|
# | chrX | 584,563 | 585,326 | 66 | 585078 | 585337 | NM | 0 | + |
# | chrX | 1,510,501 | 1,511,838 | 173 | 1510791 | 1511039 | NM | 0 | - |
# | chrX | 2,846,195 | 2,847,511 | 92 | 2847272 | 2847416 | NM | 0 | - |
# | chrX | 13,587,648 | 13,588,221 | 49 | 13587693 | 13588054 | NM | 0 | + |
# | ... | ... | ... | ... | ... | ... | ... | ... | ... |
# | chrX | 153,068,787 | 153,070,353 | 134 | 153067622 | 153070355 | NM | 0 | - |
# | chrX | 153,284,685 | 153,285,655 | 94 | 153284647 | 153284779 | NM | 0 | - |
# | chrX | 153,598,874 | 153,600,604 | 164 | 153599240 | 153599729 | NM | 0 | - |
# | chrX | 153,990,840 | 153,991,831 | 105 | 153991030 | 153991256 | NM | 0 | + |
# +--------------|-------------|-------------|-----------|------------|-----------|------------|-----------|--------------+
# Unstranded PyRanges object has 65 rows and 9 columns from 2 chromosomes.
# +--------------|-----------|-----------|-----------|------------|-----------|-----------------------------------------|-----------|-------------+
# | Chromosome | Start | End | CpG | Start_xn | End_xn | Name | Score | CpGDecile |
# | (category) | (int32) | (int32) | (int64) | (int32) | (int32) | (object) | (int64) | (float64) |
# |--------------|-----------|-----------|-----------|------------|-----------|-----------------------------------------|-----------|-------------|
# | chrX | 584563 | 585326 | 66 | 585078 | 585337 | NM_000451_exon_0_0_chrX_585079_f | 0 | 6.6 |
# | chrX | 1510501 | 1511838 | 173 | 1510791 | 1511039 | NM_001636_exon_3_0_chrX_1510792_r | 0 | 17.3 |
# | chrX | 2846195 | 2847511 | 92 | 2847272 | 2847416 | NM_001669_exon_9_0_chrX_2847273_r | 0 | 9.2 |
# | chrX | 13587648 | 13588221 | 49 | 13587693 | 13588054 | NM_001167890_exon_0_0_chrX_13587694_f | 0 | 4.9 |
# | ... | ... | ... | ... | ... | ... | ... | ... | ... |
# | chrX | 153068787 | 153070353 | 134 | 153067622 | 153070355 | NM_032512_exon_0_0_chrX_153067623_r | 0 | 13.4 |
# | chrX | 153284685 | 153285655 | 94 | 153284647 | 153284779 | NM_001025243_exon_10_0_chrX_153284648_r | 0 | 9.4 |
# | chrX | 153598874 | 153600604 | 164 | 153599240 | 153599729 | NM_001456_exon_45_0_chrX_153599241_r | 0 | 16.4 |
# | chrX | 153990840 | 153991831 | 105 | 153991030 | 153991256 | NM_001363_exon_0_0_chrX_153991031_f | 0 | 10.5 |
# +--------------|-----------|-----------|-----------|------------|-----------|-----------------------------------------|-----------|-------------+
# Unstranded PyRanges object has 58 rows and 9 columns from 1 chromosomes.
cpg_rle = cpg.to_rle(value_col="CpG") # ignore value_col for regular coverage
cpg_rle
# chrX
# ----
# +--------|---------|-------|--------|-------|---------|---------|-------|----------|-------|----------|-------+
# | Runs | 64181 | 612 | 4340 | 896 | 78656 | ... | 607 | 268069 | 389 | 135083 | 308 |
# |--------|---------|-------|--------|-------|---------|---------|-------|----------|-------|----------|-------|
# | Values | 0.0 | 62.0 | 0.0 | 100.0 | 0.0 | ... | 44.0 | 0.0 | 36.0 | 0.0 | 29.0 |
# +--------|---------|-------|--------|-------|---------|---------|-------|----------|-------|----------|-------+
# Rle of length 155246568 containing 1792 elements
#
# chrY
# ----
# +--------|---------|-------|--------|-------|---------|---------|-------|------------|-------|----------|-------+
# | Runs | 14181 | 612 | 4340 | 896 | 78656 | ... | 229 | 30440250 | 389 | 135083 | 308 |
# |--------|---------|-------|--------|-------|---------|---------|-------|------------|-------|----------|-------|
# | Values | 0.0 | 62.0 | 0.0 | 100.0 | 0.0 | ... | 25.0 | 0.0 | 36.0 | 0.0 | 29.0 |
# +--------|---------|-------|--------|-------|---------|---------|-------|------------|-------|----------|-------+
# Rle of length 59349574 containing 362 elements
# Unstranded PyRles object with 2 chromosomes.
Features
- fast (also in single-core mode)
- supports multiple cores
- memory-efficient
- featureful
- pythonic/pandastic
- supports chaining with a terse syntax
- uses Pandas DataFrames, so the whole Python data science stack works on PyRanges.
Supporting pyranges
- most importantly, cite pyranges if you use it. It is the main metric funding sources care about.
- use pyranges in Stack Overflow/biostars questions and answers
- star the repo (possibly important for github visibility and as a proxy for project popularity)
- if you are a business using pyranges, please give to one of the charities listed at https://www.givewell.org/
Documentation
https://biocore-ntnu.github.io/pyranges/
(Might be slightly out of date; watch the CHANGELOG too)
Install
The preferred way to install pyranges is through the bioconda channel:
conda install -c bioconda pyranges
You can also try pip:
pip install pyranges
PyRanges has some dependencies that are optional. They need to be manually installed if you require their functionality:
ray: multicpu # pip install -U ray
pybigwig: write bigwigs # pip install pybigwig or conda install -c bioconda pybigwig
bamread: read bam files # pip install bamread or conda install -c bioconda bamread
fisher: fast fisher exact # pip install fisher or conda install -c bioconda fisher
Since these are not needed for 99.9% percent of the pyranges functionality, they are kept separate to prevent the possibility of the pyranges-install failing due to dependencies that fail installation or conflicting dependencies.
Paper/Cite
http://dx.doi.org/10.1093/bioinformatics/btz615
TODO
For the future:
- write docstrings, autogenerate API-docs
PyRanges should always be the fastest general-purpose genomics library for Python. So I will happily change the multithreading library and overlap datastructures sometime in the future, if rigorous tests show that the proposed alternatives are indeed faster. (As the multithreading requires about 30 lines of code and the overlap queries about 15, this will not be hard.)
Performance
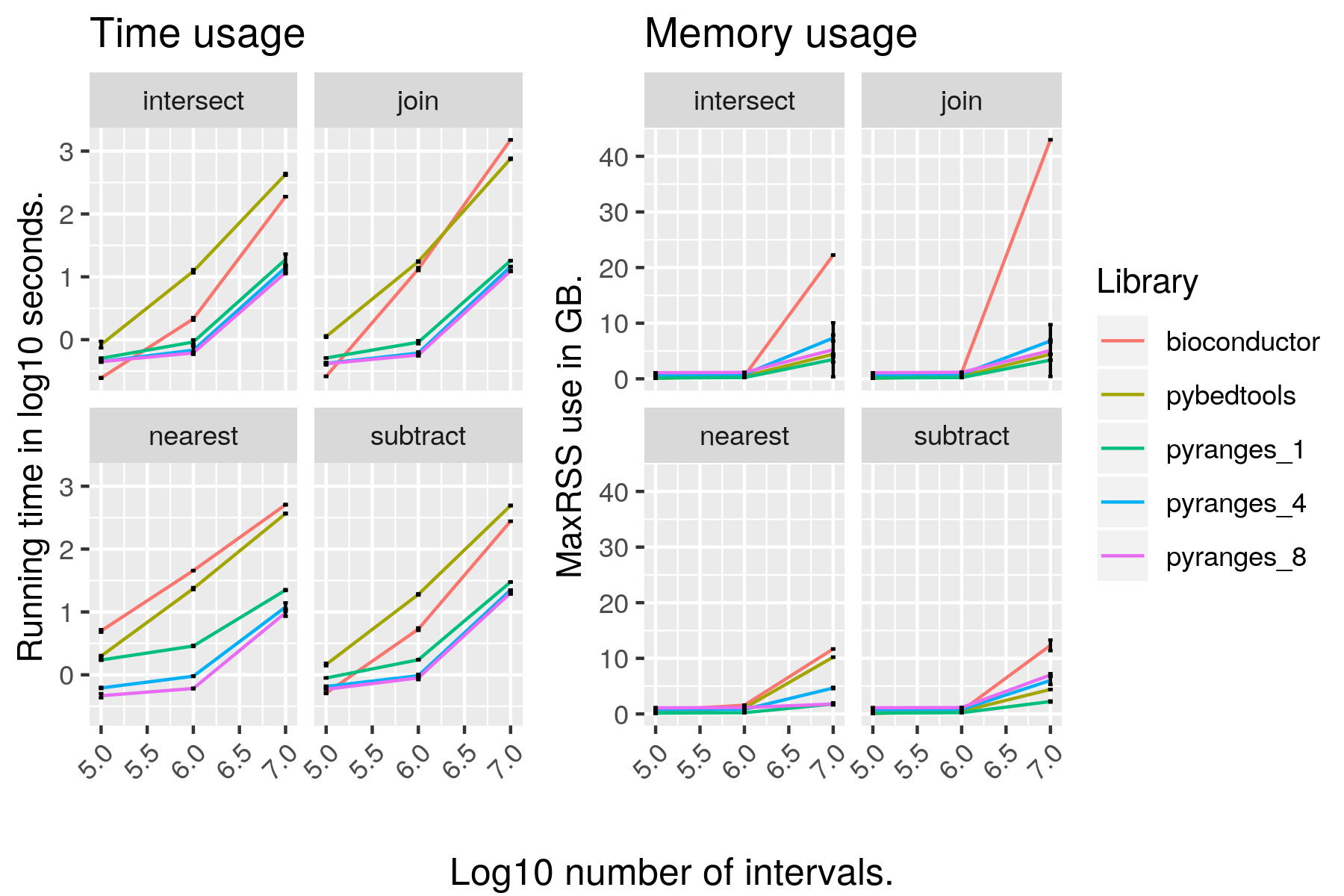
Comprehensive set of graphs for many types of functions on different datasets are here:
The exact code tested is found here.




