REPTILE
- Regulatory Element Prediction based on TIssue-specific Local Epigenetic marks
https://github.com/yupenghe/REPTILE
REPTILE is a tool to identify the precise location of enhancers by integrating histone modification data and base-resolution DNA methylation profiles.
Please contact Yupeng He for for feedbacks, questions or bugs.
Supplemental Data for REPTILE manuscript can be downloaded from here.
TABLE OF CONTENTS
- Requirement
- Installation
- Uninstallation
- Test REPTILE
- REPTILE Overview
- Use REPTILE
- Example
- File format
REQUIREMENT
R
REPTILE requires R (>= 3.2.2) and it is available in R website.
python
REPTILE requires python2 (>= 2.7.9) or python3 (>= 3.5.1). It also requires numpy (>= 1.10.4) and pandas (>= 0.17.1) modules. The numpy and pandas are available from their websties. It is recommended to install anaconda for python2.7 and the two modules will be included.
bedtools
1 - bedtools is available in bedtools website. It is recommended to download and install the latest stable release, according to the installation instruction. Older versions may work but they have not been tested.
2 - After installation, please include the path to the bedtools executable in PATH environmental variable.
The executable is usually in the bin/ folder within the bedtools/ folder. Suppose that the path of bedtools
executable is /path/to/bedtools/bin/, you can add the below command to your ~/.bashrc file (your shell config file)
to accomplish this requirement.
export PATH=/path/to/bedtools/bin/:$PATH3 - Please check whether the bedtools executable has excecute permission. If not, you will error message
"Permission denied". The command below can solve this issue.
chmod u+x /path/to/bedtools/bin/bedtoolsbigWigAverageOverBed
1 - bigWigAverageOverBed executable can be downloaded from
UCSC genome browser utilities.
Search for "bigWigAverageOverBed" under the folder of your operation system.
For example, here are the binary releases for
linux and
macOS.
2 - After bigWigAverageOverBed is downloaded, please run the below command to give it execute permission.
chmod u+x bigWigAverageOverBed3 - Last, please include the path to the bigWigAverageOverBed executable in PATH environmental variable.
If the path of executable is /path/to/bigWigAverageOverBed/, you can add the below command to ~/.bashrc
file (your shell config file) to accomplish this requirement.
export PATH=/path/to/bigWigAverageOverBed/:$PATHINSTALLATION
1 - REPTILE can be downloaded (cloned) using git command.
git clone https://github.com/yupenghe/REPTILE.git2 - The R library required for REPTILE can be installed through CRAN by command below.
R
> install.packages("REPTILE")More information about this R package is available in REPTILE CRAN webpage.
3 - To make REPTILE easier to use, it is recommended to add the path of executable scripts (in the bin/ folder)
from REPTILE in PATH environmental variable Otherwise, the path of the scripts has to be typed for each use.
Suppose the path is /path/to/REPTILE/bin/, please add the below command to ~/.bashrc file
(your shell config file).
export PATH=/path/to/REPTILE/bin/:$PATHUNINSTALLATION
To uninstall REPTILE, please run the below command to remove the installed R package. You may want to remove the REPTILE folder and related files to fully clean up.
R CMD UNINSTALL REPTILETEST REPTILE
Below command can be used to test whether REPTILE is correctly installed and all requirements are met.
cd REPTILE/test
./run_test.pyREPTILE OVERVIEW
Key concepts
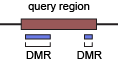
REPTILE is a tool to identify enhancers by epigenomic data (DNA methylation and histone modifications). There two key concepts in REPTILE, "query region" and "DMR":
-
query region - (training) known enhancers and regions with no observable enhancer activity. (prediction) sliding genomic windowns used to call the enhancers that show little methylation variation (and thus contain no DMR).
-
DMR - differentially methylated regions, used as high-resolution enhancer candidates
The currently known enhancers (~2kb) are generally much larger than the binding motifs of TFs (~10-20bp) and are likely to include sequences that contribute little to their enhancer activity. We used the term “query region” to describe such large regions where a small fraction of the region is regulatory (if any). Query region also refers to negative regions (that showed no observable enhancer activity) and the genomic windows used by enhancer prediction methods. Since large portion of an active query region is likely to have little contribution to its enhancer activity, the epigenomic signature of the whole active query region may not be a good approximation to the epigenomic state of the bona fide regulatory sequences within it. To address this issue, we used differentially methylated regions (DMRs, ~500bp) to pinpoint the possible regulatory sub-regions within query regions. DMRs serve as high-resolution enhancer candidates and may capture the local epigenomic patterns that would otherwise be averaged/washed out in analysis focusing on the query regions.
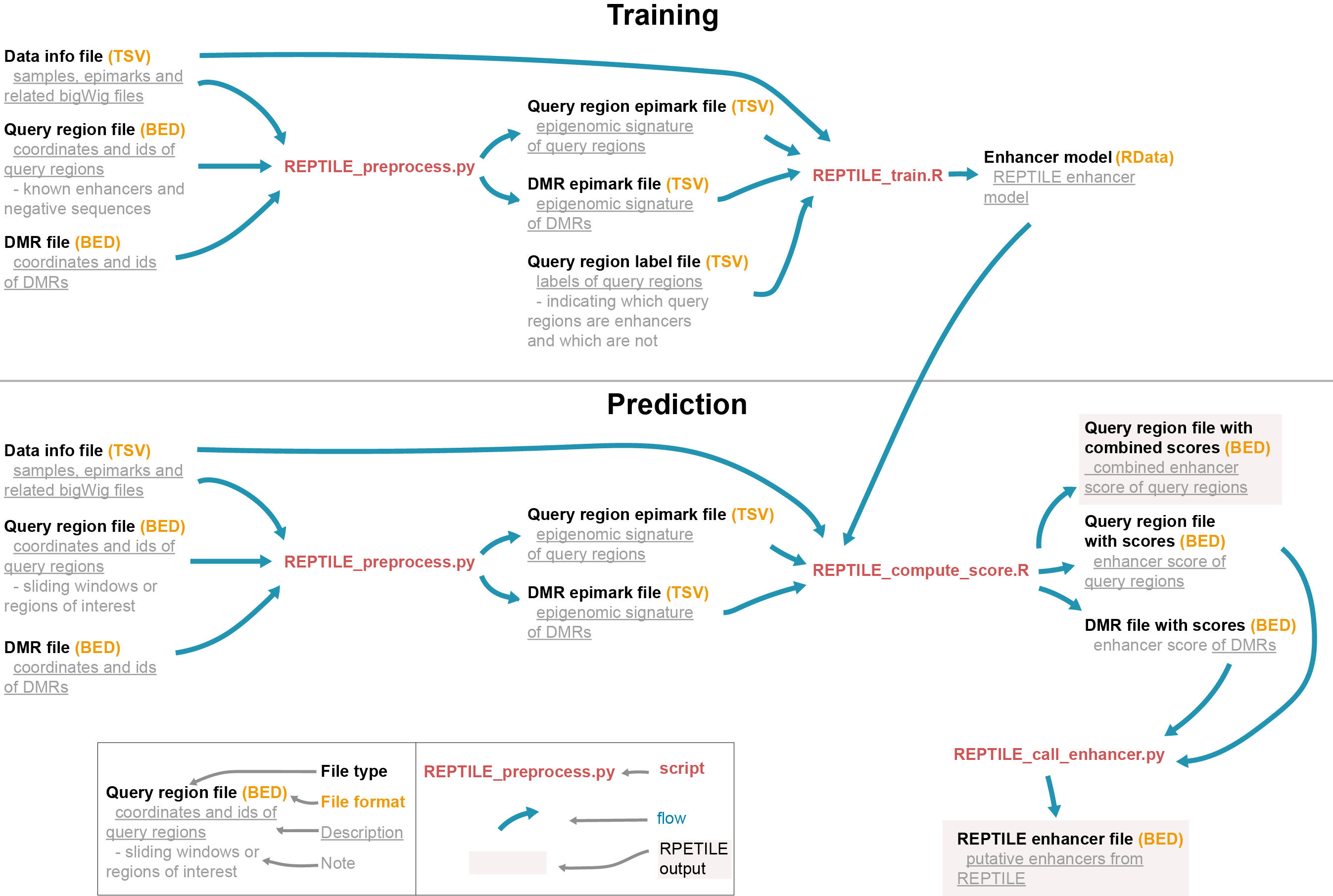
REPTILE workflow
The workflow of REPTILE pipeline:
Check File format section for details of each file.
USE REPTILE
As shown in the REPTILE workflow, REPTILE pipeline can be used to 1) learn
enhancer models from the epigenomic signature of known enhancers and negative sequences and 2)
then generate enhancer prediction based on the model. The pipeline is composed by five executable
scripts (in bin/). REPTILE_train.R and REPTILE_compute_score.R are the key scripts of doing training and
generate prediction, respectively. The inputs for these scripts can be readily generated using
REPTILE_preprocess.py. REPTILE_call_enhancer.py is used to generate enhancer calls based on the
results (enhancer scores) from REPTILE_compute_score.R. The enhancer predictions can be evaluated
using the last script, REPTILE_evaluate_prediction.R.
The whole RPETILE pipeline can be divided in the several steps. First, input files (in commonly used file formats of genomic data) are preprocessed to generate the inputs for following training and prediction. Next, in training step, REPTILE learns an enhancer model. The model will then be used to calculate enhancer scores for each query region and DMR in the prediction step. Lastly, enhancer calls will be generated based on these scores.
Calling DMRs
DMRs can improve the resolution of REPTILE predictions but DMR calling is not part of REPTILE pipeline. Several published methods are available to call DMRs, including bsseq, MOABS and DSS. The DMRs in the example data were called by methypy. It is a python module that is designed to identify DMRs across a large number of cells and tissues, whereas existing methods are better at doing pairwise comparison.
The DMR input is optional and REPTILE still generates good predictions without it. If you want to run REPTILE without DMR input, ignoring -d option when running RPETILE scripts will do.
Preprocessing
The goal of preprocessing step is to generate the files of epigenomic signatures of DMRs and query regions. These files are inputs for follow-up training and prediction steps.
REPTILE_preprocess.py \
data_info_file \
query_region_file \
-d dmr_file \
output_prefix \
-p num_processorsNote that for generating genome-wide predictions, -g option should be used. This option
should not be enabled in the preparation of inputs for training or the calculation of
enhancer scores for given regions (i.e. REPTILE_compute_score.R with -w option).
REPTILE_preprocess.py -h to get help information.
Training an enhancer model
In training step, an enhancer model is learned based on the epigenomic signatures of
known enhancers and negative sequences (query_region_epimark_file and dmr_epimark_file)
generated in preprocessing step. The file query_region_label_file specifies the labels of
query regions: which regions are enhancers and which region are negative sequencers.
REPTILE_train.R \
-i data_info_file \
-a query_region_epimark_file \
-d dmr_epimark_file \
-l query_region_label_file \
-s training_sample_name \
-o output_prefixREPTILE_train.R -h to get help information.
Generate enhancer scores
Enhancer (confidence) scores are calculated for DMRs and query regions.
REPTILE_compute_score.R \
-i data_info_file \
-m enhancer_model \
-a query_region_epimark_file \
-d dmr_epimark_file \
-s prediction_sample_name \
-o output_prefixFor generating enhancer scores of given regions, option -w should be used and in
preprocessing the inputs, -g should NOT be used (in running REPTILE_preprocess.py).
If -w is used, the combined score will also be calculated. The combined score for each
query region is the maximum of scores of the query region and DMRs overlapping with it.
REPTILE_compute_score.R -h to get help information.
Get enhancer calls
Based on the enhancer scores of DMRs and query regions, putative enhancers can be generated.
REPTILE_call_enhancers.py \
query_region_file_with_score \
-d dmr_file_with_score \
-o output_file_name \
-p score_cutoffFor genome-wide prediction, it is recommend to use sliding genomic windows as query regions.
REPTILE_call_enhancers.py -h to get help information.
Evaluating enhancer prediction results
The area under the receiver operating characteristic curve (AUROC), area under the precision-recall curve (AUPR) and other metrics will be calculated for users to evaluate the prediction accuracy.
REPTILE_evaluate_prediction.R
-p query_region_file_with_combined_score \
-l query_region_label_file \
-s target_sampletarget_sample is the sample in which enhancer scores are calculated.
REPTILE_evaluate_prediction.R -h to get help information.
EXAMPLE
Please see EXAMPLE.md for details. The simple example can be run with code below. 1 Gb hard drive space and 1 Gb memory are needed to run it.
cd simple_example/
sh run_simple_example.sh > log 2> errCode below can be used to run the complete example. Please be aware that running the complete example requires large memory and check with the requirement first.
cd example/
sh run_example.sh > log 2> errPRETRAINED MODEL
Please see PRETRAINED_MODEL.md for details.
FILE FORMAT
The formats of files invovled in the REPTILE workflow are: