tylermorganwall / Skpr
Licence: gpl-3.0
Generates and evaluates D, I, A, Alias, E, T, G, and custom optimal designs. Supports generation and evaluation of mixture and split/split-split/N-split plot designs. Includes parametric and Monte Carlo power evaluation functions. Provides a framework to evaluate power using functions provided in other packages or written by the user.
Stars: ✭ 80
Programming Languages
r
7636 projects
Projects that are alternatives of or similar to Skpr
Structural Equation Modeling For Psychologists
Lesson files used in the Structural Equation Modeling for Psychologists.
Stars: ✭ 69 (-13.75%)
Mutual labels: rstats
Darkstudio
darkstudio. A dark grey alternative to RStudio's default dark theme.
Stars: ✭ 75 (-6.25%)
Mutual labels: rstats
Cloudml
R interface to Google Cloud Machine Learning Engine
Stars: ✭ 66 (-17.5%)
Mutual labels: rstats
Accesscontrolhelper
AccessControlHelper for asp.net mvc and asp.net core, strategy based authorization
Stars: ✭ 71 (-11.25%)
Mutual labels: power
Lexisnexistools
📰 Working with newspaper data from 'LexisNexis'
Stars: ✭ 59 (-26.25%)
Mutual labels: rstats
Highcharts trendline
HighCharts demo of scatter plot, including a trend line
Stars: ✭ 79 (-1.25%)
Mutual labels: linear-regression
21 Recipes
📕 An R/rtweet edition of Matthew A. Russell's Python Twitter Recipes Book
Stars: ✭ 69 (-13.75%)
Mutual labels: rstats
Gsodr
Global Surface Summary of the Day ('GSOD') Weather Data Client for R
Stars: ✭ 72 (-10%)
Mutual labels: rstats
Freebase
👃🏽A 'usethis'-like Package for Base R Pseudo-equivalents of 'tidyverse' Code
Stars: ✭ 77 (-3.75%)
Mutual labels: rstats
Sever
🔪Good-looking problems: customise your Shiny disconnected screen and error messages
Stars: ✭ 60 (-25%)
Mutual labels: rstats
Feddata
Functions to Automate Downloading Geospatial Data Available from Several Federated Data Sources
Stars: ✭ 70 (-12.5%)
Mutual labels: rstats
Awesome Blogdown
An awesome curated list of blogs built using blogdown
Stars: ✭ 80 (+0%)
Mutual labels: rstats
Lodown
locally download and prepare publicly-available microdata
Stars: ✭ 79 (-1.25%)
Mutual labels: rstats
skpr 
Overview
skpr is an open source design of experiments suite for generating and evaluating optimal designs in R. Here is a sampling of what skpr offers:
- Generates and evaluates D, I, A, Alias, E, T, and G optimal designs, as well as user-defined custom optimality criteria.
- Supports generation and evaluation of split/split-split/…/N-split plot designs.
- Includes parametric and Monte Carlo power evaluation functions, and supports calculating power for censored responses.
- Provides an extensible framework for the user to evaluate Monte Carlo power using their own libraries.
- Includes a Shiny graphical user interface, skprGUI, that auto-generates the R code used to create and evaluate the design to improve ease-of-use and enhance reproducibility.
Installation
# To install:
install.packages("skpr")
# To install the latest version from Github:
# install.packages("devtools")
devtools::install_github("tylermorganwall/skpr")
Functions
-
gen_design()generates optimal designs from a candidate set, given a model and the desired number of runs. -
eval_design()evaluates power parametrically for linear models, for normal and split-plot designs. -
eval_design_mc()evaluates power with a Monte Carlo simulation, for linear and generalized linear models. This function also supports calculating power for split-plot designs using REML. -
eval_design_survival_mc()evaluates power with a Monte Carlo simulation, allowing the user to specify a point at which the data is censored. -
eval_design_custom_mc()allows the user to import their own libraries and use the Monte Carlo framework provided by skpr to calculate power. -
skprGUI()andskprGUIbrowser()opens up the GUI in either R Studio or an external browser.
If addition, the package offers two functions to generate common plots related to designs:
-
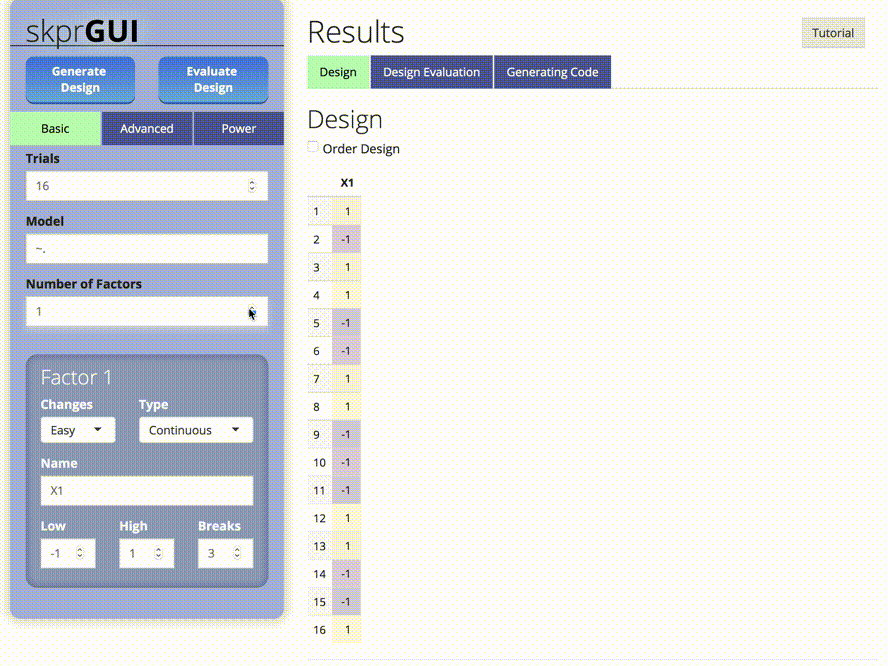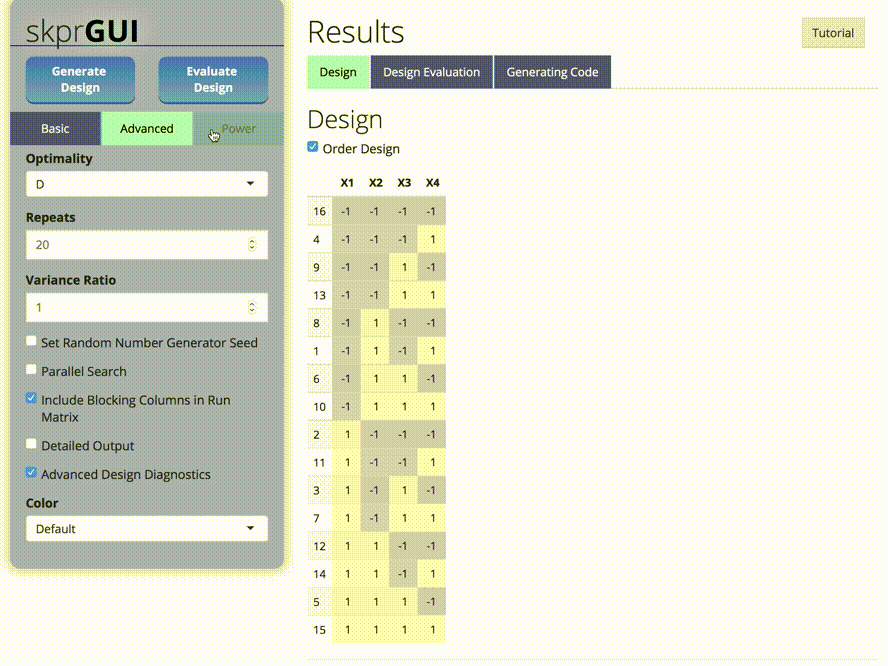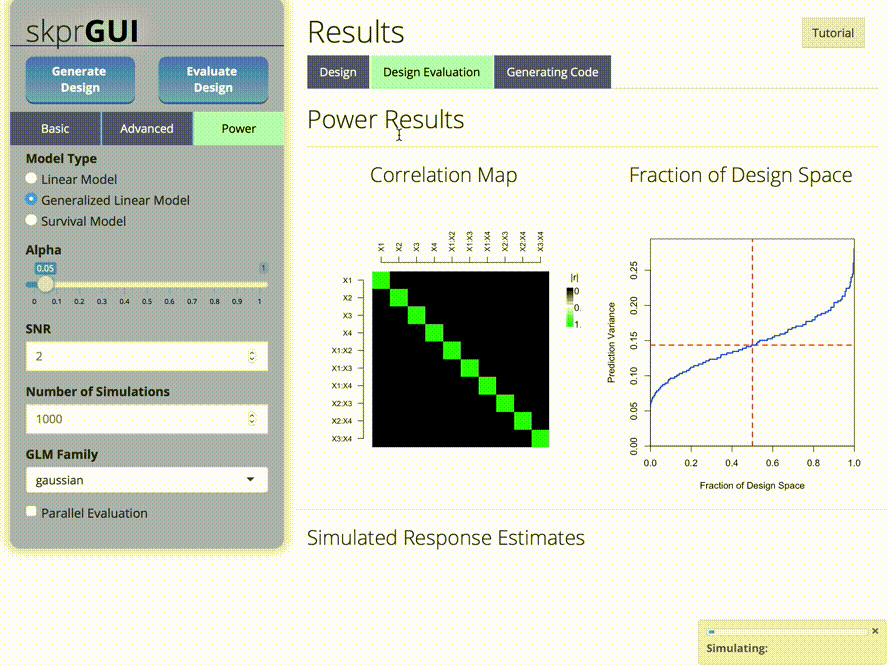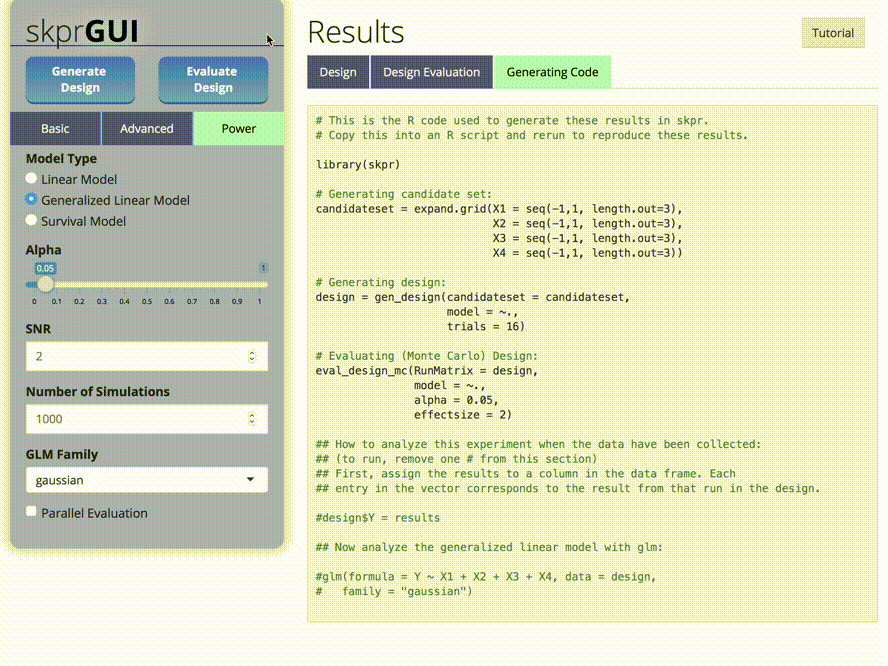
plot_correlations()generates a color map of correlations between variables. -
plot_fds()generates the fraction of design space plot for a given design.
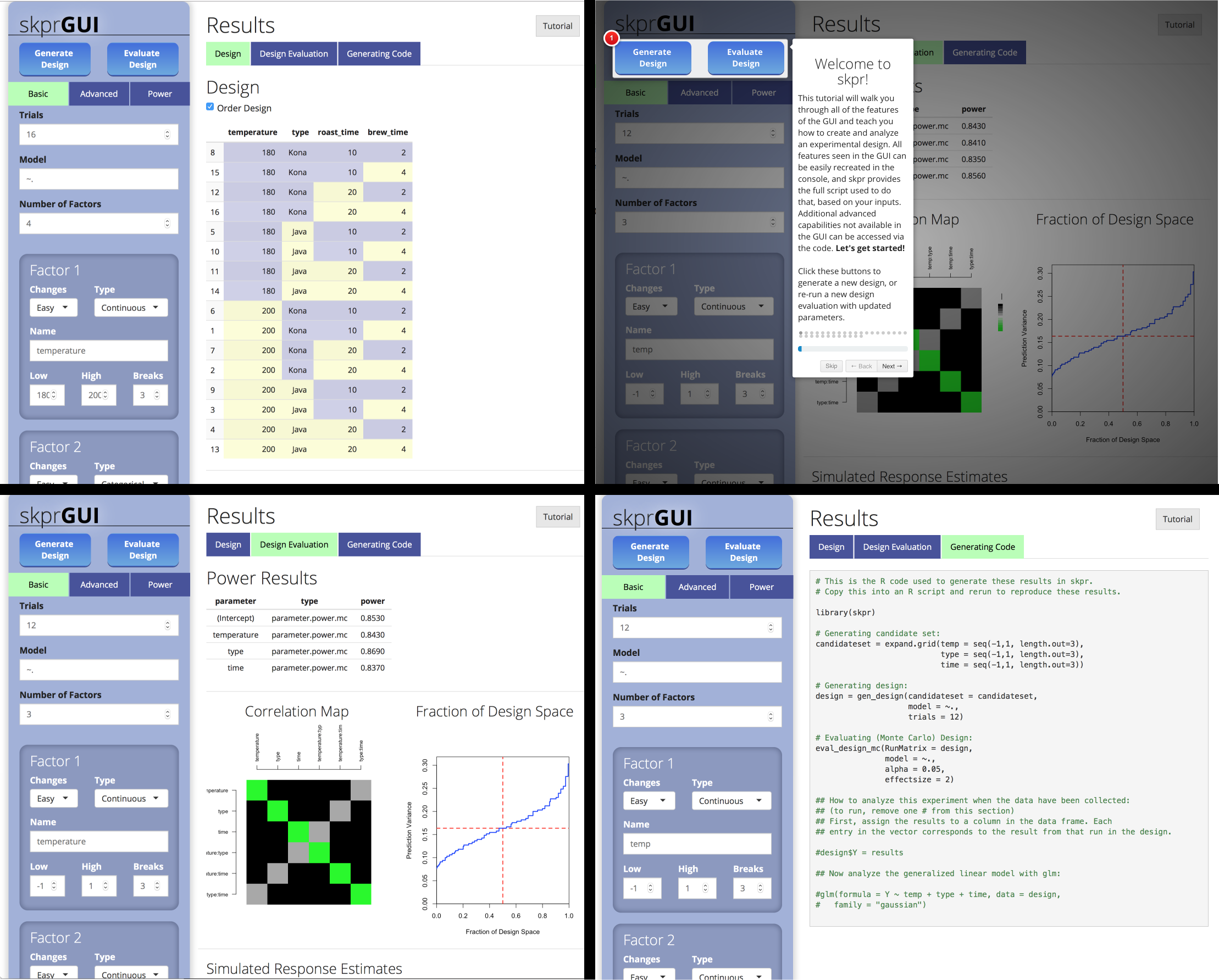
##skprGUI
skprGUI provides an graphical user interface to access all of the main
features of skpr. An interactive tutorial is provided to familiarize the
user with the available functionality. Type skprGUI() or
skprGUIbrowser() to begin. Screenshots:
Usage
library(skpr)
#Generate a candidate set of all potential design points to be considered in the experiment
#The hypothetical experiment is determining what affects the caffeine content in coffee
candidate_set = expand.grid(temp = c(80,90,100),
type = c("Kona","Java"),
beansize = c("Large","Medium","Small"))
candidate_set
#> temp type beansize
#> 1 80 Kona Large
#> 2 90 Kona Large
#> 3 100 Kona Large
#> 4 80 Java Large
#> 5 90 Java Large
#> 6 100 Java Large
#> 7 80 Kona Medium
#> 8 90 Kona Medium
#> 9 100 Kona Medium
#> 10 80 Java Medium
#> 11 90 Java Medium
#> 12 100 Java Medium
#> 13 80 Kona Small
#> 14 90 Kona Small
#> 15 100 Kona Small
#> 16 80 Java Small
#> 17 90 Java Small
#> 18 100 Java Small
#Generate the design (default D-optimal)
design = gen_design(candidateset = candidate_set,
model = ~temp + type + beansize,
trials=12)
design
#> temp type beansize
#> 1 80 Java Medium
#> 2 100 Java Large
#> 3 100 Java Small
#> 4 80 Java Large
#> 5 80 Kona Medium
#> 6 80 Kona Small
#> 7 100 Kona Small
#> 8 100 Kona Medium
#> 9 80 Kona Large
#> 10 100 Java Medium
#> 11 100 Kona Large
#> 12 80 Java Small
#Evaluate power for the design with an allowable type-I error of 5% (default)
eval_design(design)
#> parameter type power
#> 1 (Intercept) effect.power 0.8424665
#> 2 temp effect.power 0.8424665
#> 3 type effect.power 0.8424665
#> 4 beansize effect.power 0.5165386
#> 5 (Intercept) parameter.power 0.8424665
#> 6 temp parameter.power 0.8424665
#> 7 type1 parameter.power 0.8424665
#> 8 beansize1 parameter.power 0.5593966
#> 9 beansize2 parameter.power 0.5593966
#> ============Evaluation Info============
#> • Alpha = 0.05 • Trials = 12 • Blocked = FALSE
#> • Evaluating Model = ~temp + type + beansize
#> • Anticipated Coefficients = c(1.000, 1.000, 1.000, 1.000, -1.000)
#Evaluate power for the design using a Monte Carlo simulation.
#Here, we set the effect size (here, the signal-to-noise ratio) to 1.5.
eval_design_mc(design, effectsize=1.5)
#> parameter type power
#> 1 (Intercept) effect.power.mc 0.600
#> 2 temp effect.power.mc 0.612
#> 3 type effect.power.mc 0.610
#> 4 beansize effect.power.mc 0.316
#> 5 (Intercept) parameter.power.mc 0.600
#> 6 temp parameter.power.mc 0.612
#> 7 type1 parameter.power.mc 0.610
#> 8 beansize1 parameter.power.mc 0.359
#> 9 beansize2 parameter.power.mc 0.354
#> ===========Evaluation Info============
#> • Alpha = 0.05 • Trials = 12 • Blocked = FALSE
#> • Evaluating Model = ~temp + type + beansize
#> • Anticipated Coefficients = c(0.750, 0.750, 0.750, 0.750, -0.750)
#Evaluate power for the design using a Monte Carlo simulation, for a non-normal response.
#Here, we also increase the number of simululations to improve the precision of the results.
eval_design_mc(design, nsim=5000, glmfamily = "poisson", effectsize=c(2,6))
#> parameter type power
#> 1 (Intercept) effect.power.mc 0.9968
#> 2 temp effect.power.mc 0.9826
#> 3 type effect.power.mc 0.9832
#> 4 beansize effect.power.mc 0.8502
#> 5 (Intercept) parameter.power.mc 0.9968
#> 6 temp parameter.power.mc 0.9826
#> 7 type1 parameter.power.mc 0.9832
#> 8 beansize1 parameter.power.mc 0.8842
#> 9 beansize2 parameter.power.mc 0.7052
#> ============Evaluation Info============
#> • Alpha = 0.05 • Trials = 12 • Blocked = FALSE
#> • Evaluating Model = ~temp + type + beansize
#> • Anticipated Coefficients = c(1.242, 0.549, 0.549, 0.549, -0.549)
#skpr was designed to operate with the pipe (%>%) in mind.
#Here is an example of an entire design of experiments analysis in three lines:
expand.grid(temp = c(80,90,100), type = c("Kona","Java"), beansize = c("Large","Medium","Small")) %>%
gen_design(model = ~temp + type + beansize + beansize:type + I(temp^2), trials=24, optimality="I") %>%
eval_design_mc()
#> parameter type power
#> 1 (Intercept) effect.power.mc 0.912
#> 2 temp effect.power.mc 0.927
#> 3 type effect.power.mc 0.997
#> 4 beansize effect.power.mc 0.935
#> 5 I(temp^2) effect.power.mc 0.637
#> 6 type:beansize effect.power.mc 0.913
#> 7 (Intercept) parameter.power.mc 0.912
#> 8 temp parameter.power.mc 0.927
#> 9 type1 parameter.power.mc 0.997
#> 10 beansize1 parameter.power.mc 0.917
#> 11 beansize2 parameter.power.mc 0.913
#> 12 I(temp^2) parameter.power.mc 0.637
#> 13 type1:beansize1 parameter.power.mc 0.899
#> 14 type1:beansize2 parameter.power.mc 0.902
#> ==============Evaluation Info==============
#> • Alpha = 0.05 • Trials = 24 • Blocked = FALSE
#> • Evaluating Model = ~temp + type + beansize + type:beansize + I(temp^2)
#> • Anticipated Coefficients = c(1.000, 1.000, 1.000, 1.000, -1.000, 1.000, 1.000, -1.000)
Note that the project description data, including the texts, logos, images, and/or trademarks,
for each open source project belongs to its rightful owner.
If you wish to add or remove any projects, please contact us at [email protected].